7XYM
 
 | |
5IZU
 
 | |
5JXC
 
 | | SynGAP Coiled-coil trimer | | Descriptor: | Ras/Rap GTPase-activating protein SynGAP | | Authors: | Shang, Y, Zhang, M. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity.
Cell, 166, 2016
|
|
7F7G
 
 | | a linear Peptide Inhibitors in complex with GK domain | | Descriptor: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
7F7I
 
 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | Descriptor: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
5JXB
 
 | | PSD-95 extended PDZ3 in complex with SynGAP PBM | | Descriptor: | Disks large homolog 4,SynGAP | | Authors: | Shang, Y, Zhang, M. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity.
Cell, 166, 2016
|
|
4G2V
 
 | | Structure complex of LGN binding with FRMPD1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, G-protein-signaling modulator 2, ... | | Authors: | Shang, Y, Pan, Z, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2012-07-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the interaction between LGN and Frmpd1
J.Mol.Biol., 425, 2013
|
|
4DC2
 
 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
5B64
 
 | | A novel binding mode of MAGUK GK domain revealed by DLG GK domain in complex with KIF13B MBS domain | | Descriptor: | DLG GK, GLYCEROL, Protein Kif13b, ... | | Authors: | Shang, Y, Zhu, J, Zhang, M. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An Atypical MAGUK GK Target Recognition Mode Revealed by the Interaction between DLG and KIF13B
Structure, 24, 2016
|
|
3RO2
 
 | | Structures of the LGN/NuMA complex | | Descriptor: | G-protein-signaling modulator 2, GLYCEROL, peptide of Nuclear mitotic apparatus protein 1 | | Authors: | Shang, Y, Wei, Z. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
5GMO
 
 | | X-ray structure of carbonyl reductase SsCR | | Descriptor: | Protein induced by osmotic stress | | Authors: | Shang, Y.P, Yu, H.L, Xu, J.H. | | Deposit date: | 2016-07-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Efficient Synthesis of (R)-2-Chloro-1-(2,4-dichlorophenyl)ethanol with a Ketoreductase from Scheffersomyces stipitis CBS 6045
Adv.Synth.Catal., 359, 2017
|
|
5GNV
 
 | | Structure of PSD-95/MAP1A complex reveals unique target recognition mode of MAGUK GK domain | | Descriptor: | Disks large homolog 4, Microtubule-associated protein 1A, SULFATE ION | | Authors: | Shang, Y, Xia, Y, Zhu, R, Zhu, J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of the PSD-95/MAP1A complex reveals a unique target recognition mode of the MAGUK GK domain
Biochem. J., 474, 2017
|
|
3UAT
 
 | |
5YW4
 
 | | Structure-Guided Engineering of Reductase: Efficient Attenuating Substrate Inhibition in Asymmetric Catalysis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Li, A.T, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
5YWL
 
 | | SsCR_L211H | | Descriptor: | Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Li, A.T, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
5YWN
 
 | | SsCR_L211H-NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
3KEX
 
 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G6H
 
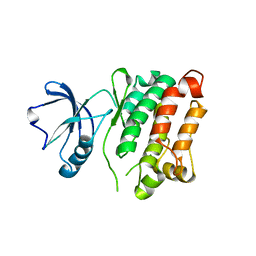 | | Src Thr338Ile inhibited in the DFG-Asp-Out conformation | | Descriptor: | N-{4-methyl-3-[(3-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
3G6G
 
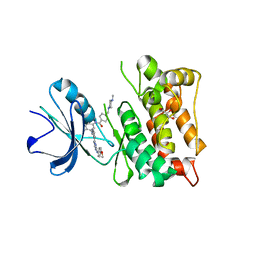 | | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations | | Descriptor: | GLYCEROL, N-{3-[(3-{4-[(4-methoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]-4-methylphenyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
2M20
 
 | | EGFR transmembrane - juxtamembrane (TM-JM) segment in bicelles: MD guided NMR refined structure. | | Descriptor: | Epidermal growth factor receptor | | Authors: | Endres, N.F, Das, R, Smith, A, Arkhipov, A, Kovacs, E, Huang, Y, Pelton, J.G, Shan, Y, Shaw, D.E, Wemmer, D.E, Groves, J.T, Kuriyan, J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor.
Cell(Cambridge,Mass.), 152, 2013
|
|
7XTK
 
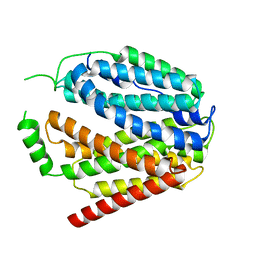 | | Cryo-EM structure of SLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, M.F, Shan, Y.Y. | | Deposit date: | 2022-05-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of human SLC19A1.
To Be Published
|
|
4F3L
 
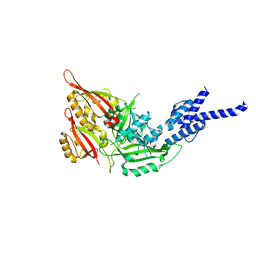 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
6FS3
 
 | | Phosphotriesterase PTE_A53_1 | | Descriptor: | (2~{S})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Bacterail Phosphotriesterase variant with high catalytic activity towards organophosphate nerve agents developed by use of structure-based design and molecular evolution
To Be Published
|
|
8P7I
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{S},6~{R})-2,6-dimethyl-1,3-dioxane-4,4-diol, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7F
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | Parathion hydrolase, ZINC ION | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
