7TTC
 
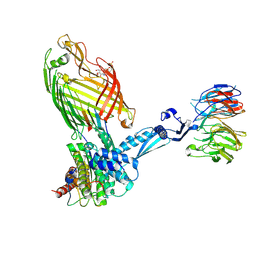 | | BamABCDE bound to substrate EspP | | Descriptor: | 2,3-dideoxy-6-O-[2-deoxy-4-O-phosphono-2-(tetradecanoylamino)-alpha-L-gulopyranosyl]-1-O-phosphono-beta-D-threo-hexopyranose, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT5
 
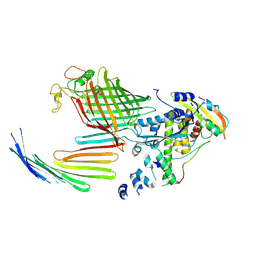 | | BamABCDE bound to substrate EspP in the open-sheet EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT2
 
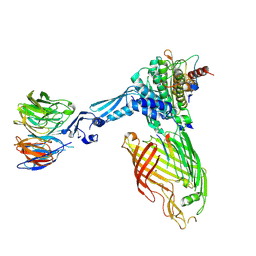 | | BamABCDE bound to substrate EspP class 3 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
2XZ1
 
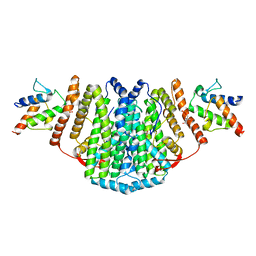 | | The Structure of the 2:2 (Fully Occupied) Complex Between Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) and Acyl Carrier Protein. | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACYL CARRIER PROTEIN 1, CHLOROPLASTIC, ... | | Authors: | Guy, J.E, Moche, M, Whittle, E, Lengqvist, J, Shanklin, J, Lindqvist, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Remote Control of Regioselectivity in Acyl-Acyl Carrier Protein-Desaturases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZYS
 
 | | Human dynamin 1 deltaPRD polymer stabilized with GMPPCP | | Descriptor: | DYNAMIN-1, INTERFERON-INDUCED GTP-BINDING PROTEIN MX1 | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
3ZBX
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor 6-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl) quinoline. | | Descriptor: | 6-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J, Ryan, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
5W0B
 
 | | Structure of human TUT7 catalytic module (CM) | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1Z71
 
 | | thrombin and P2 pyridine N-oxide inhibitor complex structure | | Descriptor: | Hirudin IIIB', L17, thrombin | | Authors: | Nantermet, P.G, Burgey, C.S, Robinson, K.A, Pellicore, J.M, Newton, C.L, Deng, J.Z, Lyle, T.A, Selnick, H.G, Lewis, S.D, Lucas, B.J, Krueger, J.A, Miller-Stein, C, White, R.B, Wong, B, McMasters, D.R, Wallace, A.A, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | P(2) pyridine N-oxide thrombin inhibitors: a novel peptidomimetic scaffold
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
5W0O
 
 | |
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
4ZZM
 
 | | Human ERK2 in complex with an irreversible inhibitor | | Descriptor: | 7-ethylsulfonyl-N-(oxan-4-yl)-6,8-dihydro-5H-pyrido[3,4-d]pyrimidin-2-amine, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
4ZZO
 
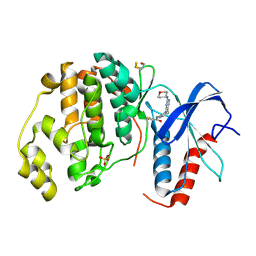 | | Human ERK2 in complex with an irreversible inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 1, N-[2-[[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
8EFS
 
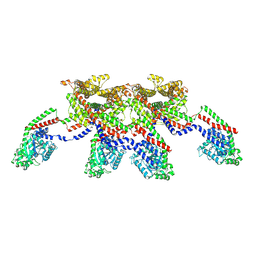 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
3ZYC
 
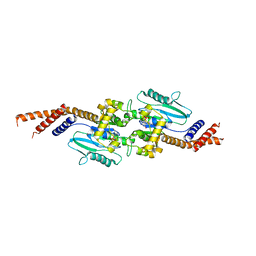 | | DYNAMIN 1 GTPASE GED FUSION DIMER COMPLEXED WITH GMPPCP | | Descriptor: | DYNAMIN-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
4ZZN
 
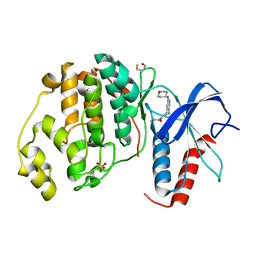 | | Human ERK2 in complex with an inhibitor | | Descriptor: | 2-[[5-chloranyl-2-(oxan-4-ylamino)pyridin-4-yl]amino]-N-methyl-benzamide, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
8F6T
 
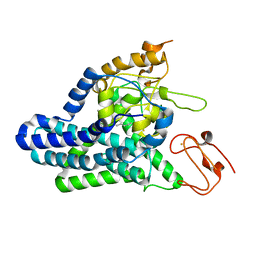 | | Cryo-EM structure of alkane 1-monooxygenase AlkB-AlkG complex from Fontimonas thermophila | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION | | Authors: | Chai, J, Guo, G, McSweeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for enzymatic terminal C-H bond functionalization of alkanes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EF7
 
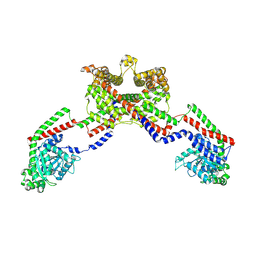 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
4CXP
 
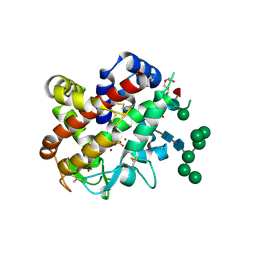 | | Structure of bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana in complex with sulfate | | Descriptor: | ENDONUCLEASE 2, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CWM
 
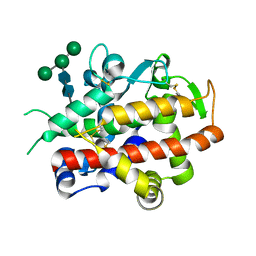 | | High-glycosylation crystal structure of the bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ZINC ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXO
 
 | | bifunctional endonuclease in complex with ssDNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ENDONUCLEASE 2, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4CXV
 
 | | Structure of bifunctional endonuclease (AtBFN2) in complex with phosphate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
1NJS
 
 | | human GAR Tfase in complex with hydrolyzed form of 10-trifluoroacetyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Marsilje, T.H, Li, C, Hedrick, M.P, Gooljarsingh, L.T, Tavassoli, A, Benkovic, S.J, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational Design, Synthesis, Evaluation, and Crystal Structure of a Potent Inhibitor of Human GAR Tfase: 10-(Trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic Acid
Biochemistry, 42, 2003
|
|
1BBC
 
 | | STRUCTURE OF RECOMBINANT HUMAN RHEUMATOID ARTHRITIC SYNOVIAL FLUID PHOSPHOLIPASE A2 AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Wery, J.-P, Schevitz, R.W, Clawson, D.K, Bobbitt, J.L, Dow, E.R, Gamboa, G, Goodsonjunior, T, Hermann, R.B, Kramer, R.M, Mcclure, D.B, Mihelich, E.D, Putnam, J.E, Sharp, J.D, Stark, D.H, Teater, C, Warrick, M.W, Jones, N.D. | | Deposit date: | 1992-05-04 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant human rheumatoid arthritic synovial fluid phospholipase A2 at 2.2 A resolution.
Nature, 352, 1991
|
|
5W0N
 
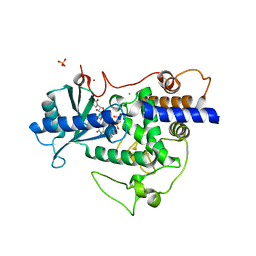 | | Structure of human TUT7 catalytic module (CM) in complex with UMPNPP and U2 RNA | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3FPR
 
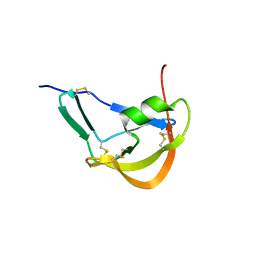 | | Crystal Structure of Evasin-1 | | Descriptor: | Evasin-1 | | Authors: | Dias, J.M, Shaw, J.P. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Plos One, 4, 2009
|
|
