1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
2FXT
 
 | | Crystal Structure of Yeast Tim44 | | Descriptor: | Import inner membrane translocase subunit TIM44 | | Authors: | Josyula, R, Sha, B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Yeast Mitochondrial Peripheral Membrane Protein Tim44p C-terminal Domain.
J.Mol.Biol., 359, 2006
|
|
1MO0
 
 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
2GW1
 
 | | Crystal Structure of the Yeast Tom70 | | Descriptor: | Mitochondrial precursor proteins import receptor | | Authors: | Wu, Y, Sha, B. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2QLD
 
 | | human Hsp40 Hdj1 | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Hu, J, Wu, Y, Li, J, Fu, Z, Sha, B. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1.
Bmc Struct.Biol., 8, 2008
|
|
3QFP
 
 | | Crystal structure of yeast Hsp70 (Bip/Kar2) ATPase domain | | Descriptor: | 78 kDa glucose-regulated protein homolog, PHOSPHATE ION | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3FP2
 
 | | Crystal structure of Tom71 complexed with Hsp82 C-terminal fragment | | Descriptor: | ATP-dependent molecular chaperone HSP82, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP3
 
 | | Crystal structure of Tom71 | | Descriptor: | CHLORIDE ION, SULFATE ION, TPR repeat-containing protein YHR117W | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP4
 
 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3QFU
 
 | | Crystal structure of Yeast Hsp70 (Bip/kar2) complexed with ADP | | Descriptor: | 78 kDa glucose-regulated protein homolog, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QLE
 
 | | Structural Basis for the Function of Tim50 in the Mitochondrial Presequence Translocase | | Descriptor: | ACETATE ION, CALCIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Qian, X.G, Gebert, M, Hpker, J, Yan, M, Li, J.Z, Wiedemann, N, Laan, M.V.D, Pfanner, N, Sha, B.D. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Structural basis for the function of tim50 in the mitochondrial presequence translocase.
J.Mol.Biol., 411, 2011
|
|
3QML
 
 | | The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor | | Descriptor: | 78 kDa glucose-regulated protein homolog, MAGNESIUM ION, Nucleotide exchange factor SIL1, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-02-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QK9
 
 | | Yeast Tim44 C-terminal domain complexed with Cymal-3 | | Descriptor: | CHLORIDE ION, Mitochondrial import inner membrane translocase subunit TIM44 | | Authors: | Cui, W, Josyula, R, Fu, Z, Sha, B. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Membrane Binding Mechanism of Yeast Mitochondrial Peripheral Membrane Protein TIM44.
Protein Pept.Lett., 18, 2011
|
|
5SV7
 
 | | The Crystal structure of a chaperone | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7CAY
 
 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7L83
 
 | | NMR solution structure of Nav1.5 DIV S3b-S4a paddle motif in DPC micelle | | Descriptor: | Sodium channel protein type 5 subunit alpha | | Authors: | Hussein, A.K, Bhuiyan, M.H, Arshava, B, Zhuang, J, Poget, S.F. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and analysis of isolated S3b-S4a motif of repeat IV of the human cardiac sodium channel
Biorxiv, 2021
|
|
5ZWS
 
 | |
4MCN
 
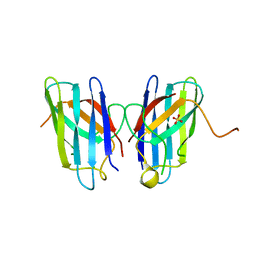 | | Human SOD1 C57S Mutant, Metal-free | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sea, K, Sohn, S.H, Durazo, A, Sheng, Y, Shaw, B, Cao, X, Taylor, A.B, Whitson, L.J, Holloway, S.P, Hart, P.J, Cabelli, D.E, Gralla, E.B, Valentine, J.S. | | Deposit date: | 2013-08-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the role of the unusual disulfide bond in copper-zinc superoxide dismutase.
J.Biol.Chem., 290, 2015
|
|
6XI6
 
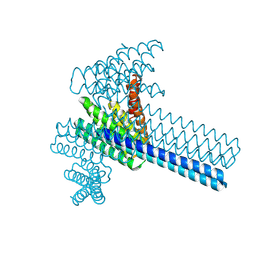 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | Descriptor: | helical fusion design | | Authors: | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
7AJR
 
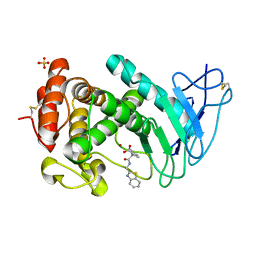 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8DC2
 
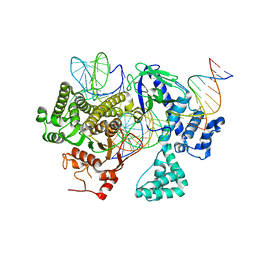 | | Cryo-EM structure of CasLambda (Cas12l) bound to crRNA and DNA | | Descriptor: | CasLambda, DNA NTS, DNA TS, ... | | Authors: | Al-Shayeb, B, Skopintsev, P, Soczek, K, Doudna, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors.
Cell, 185, 2022
|
|
4RG5
 
 | | Crystal Structure of S. Pombe SMN YG-Dimer | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein, Survival Motor Neuron protein chimera, ... | | Authors: | Gupta, K, Martin, R.S, Sarachan, K.L, Sharp, B, Van Duyne, G.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomeric Properties of Survival Motor NeuronGemin2 Complexes.
J.Biol.Chem., 290, 2015
|
|
5ZF6
 
 | | Crystal structure of the dimeric human PNPase | | Descriptor: | Polyribonucleotide nucleotidyltransferase 1, mitochondrial | | Authors: | Yuan, H.S, Golzarroshan, B. | | Deposit date: | 2018-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities.
Nucleic Acids Res., 46, 2018
|
|
7ZCK
 
 | | Room temperature crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Phosphonate ABC type transporter/ substrate binding component | | Authors: | Mikolajek, H, Shah, B.S, Paulsen, I.T, Sandy, J, Sanchez-Weatherby, J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
2L87
 
 | | The 27-residue N-terminus CCR5-peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Schnur, E, Noah, E, Ayzenshtat, I, Sargsyan, H, Inui, T, Ding, F.X, Arshava, B, Sagi, Y, Kessler, N, Levy, R, Scherf, T, Naider, F, Anglister, J. | | Deposit date: | 2011-01-06 | | Release date: | 2011-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Conformation and Orientation of a 27-Residue CCR5 Peptide in a Ternary Complex with HIV-1 gp120 and a CD4-Mimic Peptide.
J.Mol.Biol., 410, 2011
|
|
