6I04
 
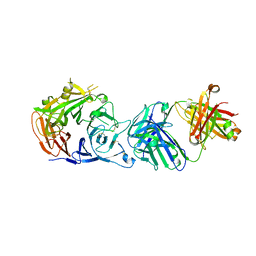 | | Crystal structure of Sema domain of the Met receptor in complex with FAB | | Descriptor: | Fab heavy chain, Fab light chain, Hepatocyte growth factor receptor | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I07
 
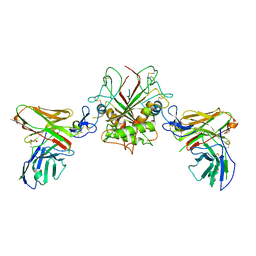 | | Crystal structure of EpCAM in complex with scFv | | Descriptor: | Epithelial cell adhesion molecule, GLYCEROL, Single chain Fv | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HYG
 
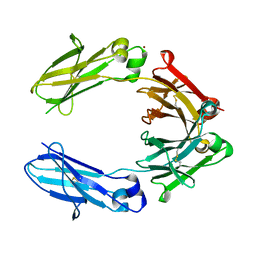 | | Heteromeric tandem IgG4/IgG1 Fc | | Descriptor: | IgHG1 and IgHG4 hybrid, ZINC ION | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S77
 
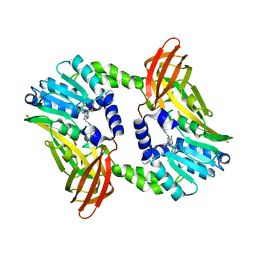 | | Crystal structure of CARM1 N265Y mutant in complex with inhibitor AA183 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-(pyridin-2-ylamino)propyl]amino]-2-azanyl-butanoic acid, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S7C
 
 | | Crystal structure of CARM1 in complex with inhibitor UM079 | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]propyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S71
 
 | | Crystal structure of CARM1 in complex with inhibitor WH5C | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(5-carbamimidamidopentyl)amino]-2-azanyl-butanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S7B
 
 | | Crystal structure of CARM1 in complex with inhibitor UM249 | | Descriptor: | 1-[4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]butyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S70
 
 | | Crystal structure of CARM1 in complex with inhibitor UM251 | | Descriptor: | 1-[5-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]pentyl]guanidine, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S74
 
 | | Crystal structure of CARM1 in complex with inhibitor UM305 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-azanylpropyl-[3-(pyrimidin-2-ylamino)propyl]amino]methyl]oxolane-3,4-diol, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
1S88
 
 | | NMR structure of a DNA duplex with two INA nucleotides inserted opposite each other, dCTCAACXCAAGCT:dAGCTTGXGTTGAG | | Descriptor: | 5'-D(*AP*GP*CP*TP*TP*GP*(2DM)P*GP*TP*TP*GP*AP*G)-3', 5'-D(*CP*TP*CP*AP*AP*CP*(2DM)P*CP*AP*AP*GP*CP*T)-3' | | Authors: | Nielsen, C.B, Petersen, M, Pedersen, E.B, Hansen, P.E, Christensen, U.B. | | Deposit date: | 2004-01-31 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid.
Bioconjug.Chem., 15, 2004
|
|
2XHN
 
 | | Rhamnogalacturonan lyase from Aspergillus aculeatus K150A active site mutant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, RHAMNOGALACTURONASE B, ... | | Authors: | Jensen, M.H, Otten, H, Christensen, U, Borchert, T.V, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Biochemical Studies Elucidate the Mechanism of Rhamnogalacturonan Lyase from Aspergillus Aculeatus.
J.Mol.Biol., 404, 2010
|
|
3L0N
 
 | | Human orotidyl-5'-monophosphate decarboxylase in complex with 6-mercapto-UMP | | Descriptor: | 6-sulfanyluridine-5'-phosphate, Uridine 5'-monophosphate synthase | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
5A09
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-[(4R)-4-(4-cyanophenyl)-5-ethanoyl-6-methyl-2-oxidanylidene-1-[3-(trifluoromethyl)phenyl]-4H-pyrimidin-3-yl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUTROPHIL ELASTASE, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
3L0K
 
 | |
5A0B
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | (4R)-4-(4-cyanophenyl)-6-methyl-2-oxidanylidene-3-[2-oxidanylidene-2-(4-propan-2-ylpiperazin-1-yl)ethyl]-1-[3-(trifluoromethyl)phenyl]-4H-pyrimidine-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5A0A
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-[(6R)-5-ethanoyl-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidin-6-yl]pyridine-2-carbonitrile, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5A0C
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | (6S)-6-(4-cyano-2-methylsulfonyl-phenyl)-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-5-carbonitrile, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
1CPY
 
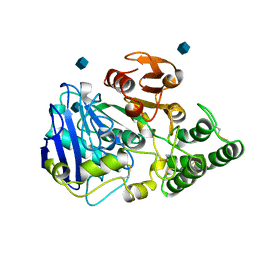 | | SITE-DIRECTED MUTAGENESIS ON (SERINE) CARBOXYPEPTIDASE Y FROM YEAST. THE SIGNIFICANCE OF THR 60 AND MET 398 IN HYDROLYSIS AND AMINOLYSIS REACTIONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE | | Authors: | Sorensen, S.B, Raaschou-Nielsen, M, Mortensen, U, Remington, S.J, Breddam, K. | | Deposit date: | 1995-03-24 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Site-Directed Mutagenesis on (Serine) Carboxypeptidase Y from Yeast. The Significance of Thr 60 and met 398 in Hydrolysis and Aminolysis Reactions
J.Am.Chem.Soc., 117, 1995
|
|
1XS4
 
 | | dCTP deaminase from Escherichia coli- E138A mutant enzyme in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
1XS1
 
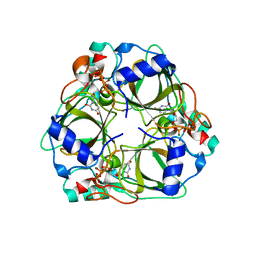 | | dCTP deaminase from Escherichia coli in complex with dUTP | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
1XS6
 
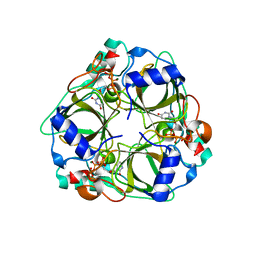 | | dCTP deaminase from Escherichia coli. E138A mutant enzyme in complex with dUTP | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
2MHZ
 
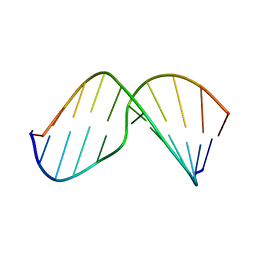 | | Structure of Exocyclic S,S N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(SDE)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2YOA
 
 | | Synaptotagmin-1 C2B domain with phosphoserine | | Descriptor: | CALCIUM ION, PHOSPHOSERINE, SYNAPTOTAGMIN-1, ... | | Authors: | Honigmann, A, van den Bogaart, G, Iraheta, E, Risselada, H.J, Milovanovic, D, Mueller, V, Muellar, S, Diederichsen, U, Fasshauer, D, Grubmuller, H, Hell, S.W, Eggeling, C, Kuhnel, K, Jahn, R. | | Deposit date: | 2012-10-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Clusters Act as Molecular Beacons for Vesicle Recruitment
Nat.Struct.Mol.Biol., 20, 2013
|
|
1UNO
 
 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
