9BPK
 
 | |
9BPI
 
 | |
9BQ5
 
 | |
9BPJ
 
 | |
1RW4
 
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
2C8V
 
 | | Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Sen, S, Krishnakumar, A, McClead, J, Johnson, M.K, Seefeldt, L.C, Szilagyi, R.K, Peters, J.W. | | Deposit date: | 2005-12-08 | | Release date: | 2006-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Role of Nucleotide-Dependent Conformational Change in Nitrogenase Catalysis: Structural Characterization of the Nitrogenase Fe Protein Leu127 Deletion Variant with Bound Mgatp.
J.Inorg.Biochem., 100, 2006
|
|
2B46
 
 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in substrate bound state | | Descriptor: | Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
2B45
 
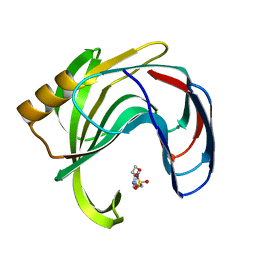 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in free state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
1W69
 
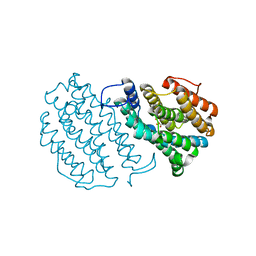 | | Crystal Structure of Mouse Ribonucleotide Reductase Subunit R2 under Reducing Conditions. A Fully Occupied Dinuclear Iron Cluster and Bound Acetate. | | Descriptor: | ACETIC ACID, FE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE M2 CHAIN | | Authors: | Karlsen, S, Strand, K.R, Kolberg, M, Rohr, A.K, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2004-08-16 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structural Studies of Changes in the Native Dinuclear Iron Center of Ribonucleotide Reductase Protein R2 from Mouse
J.Biol.Chem., 279, 2004
|
|
1W68
 
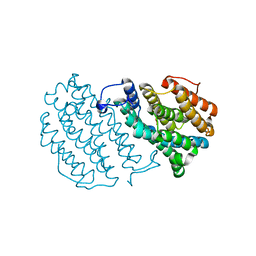 | | Crystal Structure of Mouse Ribonucleotide Reductase Subunit R2 under Oxidizing Conditions. A Fully Occupied Dinuclear Iron Cluster. | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE M2 CHAIN | | Authors: | Karlsen, S, Strand, K.R, Kolberg, M, Rohr, A.K, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2004-08-16 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structural Studies of Changes in the Native Dinuclear Iron Center of Ribonucleotide Reductase Protein R2 from Mouse
J.Biol.Chem., 279, 2004
|
|
5NM0
 
 | | Nb36 Ser85Cys with Hg, crystal form 1 | | Descriptor: | MERCURY (II) ION, Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
9GQO
 
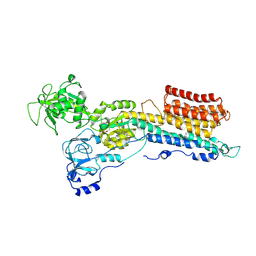 | | Structure of a Ca2+ bound phosphoenzyme intermediate in the inward-to-outward transition of Ca2+-ATPase 1 from Listeria monocytogenes | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase lmo0841, MAGNESIUM ION | | Authors: | Hansen, S.B, Flygaard, R.F, Kjaergaard, M, Nissen, P. | | Deposit date: | 2024-09-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the [Ca]E2P intermediate of Ca 2+ -ATPase 1 from Listeria monocytogenes.
Embo Rep., 26, 2025
|
|
5NLU
 
 | | Structure of Nb36 crystal form 1 | | Descriptor: | SULFATE ION, single domain llama antibody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NLW
 
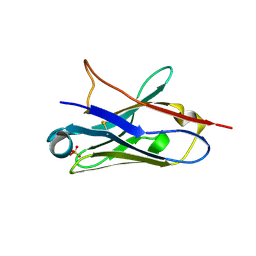 | | Structure of Nb36 crystal form 2 | | Descriptor: | SULFATE ION, nanobody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4APD
 
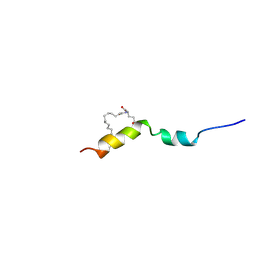 | | Liraglutide | | Descriptor: | LIRAGLUTIDE, N-hexadecanoyl-L-glutamic acid | | Authors: | Ludvigsen, S, Steensgaard, D.B, Thomsen, J.K, Strauss, H, Normann, M. | | Deposit date: | 2012-04-02 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Liraglutide
To be Published
|
|
8FW7
 
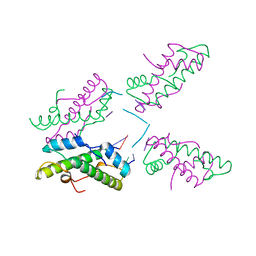 | | Histone from Bdellovibrio bacteriovorus bound to dsDNA | | Descriptor: | CBFD_NFYB_HMF domain-containing protein, DNA (5'-D(P*AP*T)-3'), DNA (5'-D(P*CP*AP*T)-3') | | Authors: | Laursen, S.P, Luger, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Histones with an unconventional DNA-binding mode in vitro are major chromatin constituents in the bacterium Bdellovibrio bacteriovorus.
Nat Microbiol, 8, 2023
|
|
8FVX
 
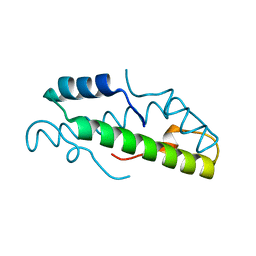 | | Histone from Bdellovibrio bacteriovorus | | Descriptor: | CBFD_NFYB_HMF domain-containing protein | | Authors: | Laursen, S.P, Luger, K. | | Deposit date: | 2023-01-19 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Histones with an unconventional DNA-binding mode in vitro are major chromatin constituents in the bacterium Bdellovibrio bacteriovorus.
Nat Microbiol, 8, 2023
|
|
5NML
 
 | | Nb36 Ser85Cys with Hg bound | | Descriptor: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1CPY
 
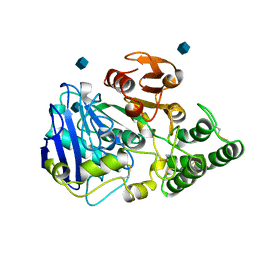 | | SITE-DIRECTED MUTAGENESIS ON (SERINE) CARBOXYPEPTIDASE Y FROM YEAST. THE SIGNIFICANCE OF THR 60 AND MET 398 IN HYDROLYSIS AND AMINOLYSIS REACTIONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE | | Authors: | Sorensen, S.B, Raaschou-Nielsen, M, Mortensen, U, Remington, S.J, Breddam, K. | | Deposit date: | 1995-03-24 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Site-Directed Mutagenesis on (Serine) Carboxypeptidase Y from Yeast. The Significance of Thr 60 and met 398 in Hydrolysis and Aminolysis Reactions
J.Am.Chem.Soc., 117, 1995
|
|
2R4R
 
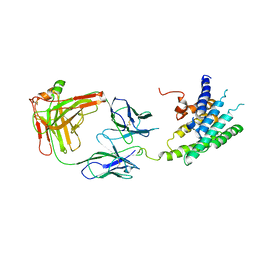 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
6ZPN
 
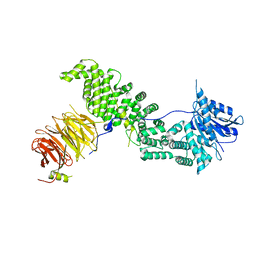 | | Crystal structure of Chaetomium thermophilum Raptor | | Descriptor: | WD_REPEATS_REGION domain-containing protein | | Authors: | Imseng, S, Boehm, R, Jakob, R.P, Hall, M.N, Hiller, S, Maier, T. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1.
Mol.Cell, 81, 2021
|
|
1BXN
 
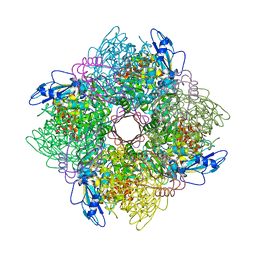 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
1HLS
 
 | | NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) | | Descriptor: | INSULIN | | Authors: | Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1995-06-28 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an engineered biologically potent insulin monomer, B16 Tyr-->His, as determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
4Y5I
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 126B with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Microtubule-associated protein tau | | Authors: | Leysen, S, Bartel, M, Milroy, L, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8FY0
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
