7XLF
 
 | | Crystal structure of CH3-THF complex of methylenetetrahydrofolate reductase from Sphingobium sp. SYK-6 | | 分子名称: | 5,10-methylenetetrahydrofolate reductase, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Yu, H.Y, Senda, M, Senda, T. | | 登録日 | 2022-04-21 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structural of methylenetetrahydrofolate reductase from Sphingobium sp. SYK-6
To Be Published
|
|
7XG9
 
 | |
3B7T
 
 | | [E296Q]LTA4H in complex with Arg-Ala-Arg substrate | | 分子名称: | IMIDAZOLE, Leukotriene A-4 hydrolase, RAR peptide, ... | | 著者 | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | 登録日 | 2007-10-31 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
1CPY
 
 | | SITE-DIRECTED MUTAGENESIS ON (SERINE) CARBOXYPEPTIDASE Y FROM YEAST. THE SIGNIFICANCE OF THR 60 AND MET 398 IN HYDROLYSIS AND AMINOLYSIS REACTIONS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE | | 著者 | Sorensen, S.B, Raaschou-Nielsen, M, Mortensen, U, Remington, S.J, Breddam, K. | | 登録日 | 1995-03-24 | | 公開日 | 1995-09-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Site-Directed Mutagenesis on (Serine) Carboxypeptidase Y from Yeast. The Significance of Thr 60 and met 398 in Hydrolysis and Aminolysis Reactions
J.Am.Chem.Soc., 117, 1995
|
|
3TRP
 
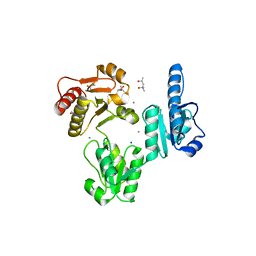 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | 著者 | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | 登録日 | 2011-09-09 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8817 Å) | | 主引用文献 | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
3O21
 
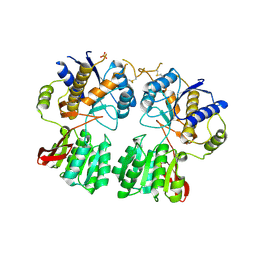 | | High resolution structure of GluA3 N-terminal domain (NTD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3, PHOSPHATE ION | | 著者 | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Babu, M.M, Jensen, M.H, Greger, I.H. | | 登録日 | 2010-07-22 | | 公開日 | 2011-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Dynamics and allosteric potential of the AMPA receptor N-terminal domain
Embo J., 30, 2011
|
|
3OBV
 
 | |
3S2I
 
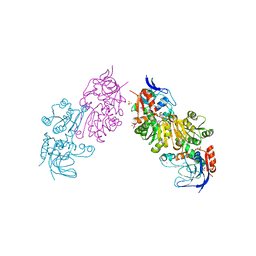 | | Crystal Structure of FurX NADH+:Furfuryl alcohol II | | 分子名称: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | 登録日 | 2011-05-16 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S2G
 
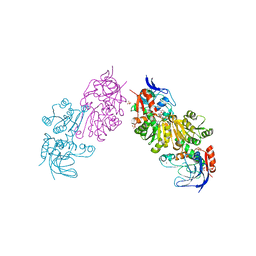 | | Crystal Structure of FurX NADH+:Furfuryl alcohol I | | 分子名称: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | 登録日 | 2011-05-16 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3ANU
 
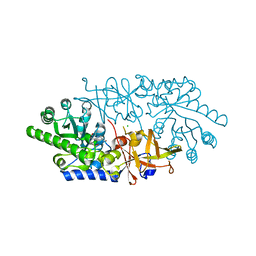 | | Crystal structure of D-serine dehydratase from chicken kidney | | 分子名称: | CHLORIDE ION, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | 登録日 | 2010-09-09 | | 公開日 | 2011-06-15 | | 最終更新日 | 2013-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
8DAD
 
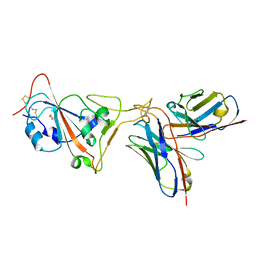 | | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AZ090 Fab Heavy Chain, AZ090 Fab Light Chain, ... | | 著者 | Zong, S, Wang, Z, Gaebler, C, Nussenzweig, M. | | 登録日 | 2022-06-13 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab
To Be Published
|
|
7FJH
 
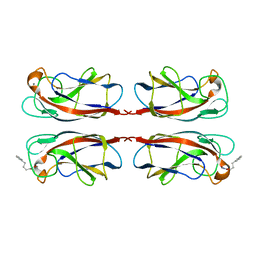 | | LecA from Pseudomonas aeruginosa in complex with 4-Phenylbutyryl hydroxamic acid (CAS: 32153-46-1) | | 分子名称: | CALCIUM ION, N-oxidanyl-4-phenyl-butanamide, PA-I galactophilic lectin | | 著者 | Shanina, S, Kuhaudomlarp, S, Siebs, E, Fuchsberger, F, Denis, M, da Silva Figueiredo Celstino Gomes, P, Clausen, M.H, Seeberger, P.H, Rognan, D, Titz, A, Imberty, A, Rademacher, C. | | 登録日 | 2021-08-04 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Targeting undruggable carbohydrate recognition sites through focused fragment library design.
Commun Chem, 5, 2022
|
|
3SEO
 
 | |
3X02
 
 | | Crystal structure of PIP4KIIBETA complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0A
 
 | | Crystal structure of PIP4KIIBETA F205L complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0B
 
 | | Crystal structure of PIP4KIIBETA I368A complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0C
 
 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X08
 
 | | Crystal structure of PIP4KIIBETA N203A complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X07
 
 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3WZZ
 
 | | Crystal structure of PIP4KIIBETA | | 分子名称: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-08 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | 分子名称: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | 分子名称: | Hepatitis C virus, ZINC ION | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-19 | | 公開日 | 2007-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
4H1V
 
 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | 分子名称: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | 登録日 | 2012-09-11 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
