2CEF
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity. | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
2CEH
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
2CFJ
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-21 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
5DVB
 
 | |
1S6Z
 
 | | Enhanced Green Fluorescent Protein Containing the Y66L Substitution | | Descriptor: | CHLORIDE ION, green fluorescent protein | | Authors: | Rosenow, M.A, Huffman, H.A, Phail, M.E, Wachter, R.M. | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Y66L Variant of Green Fluorescent Protein Supports a Cyclization-Oxidation-Dehydration Mechanism for Chromophore Maturation(,).
Biochemistry, 43, 2004
|
|
7NPA
 
 | |
6QK9
 
 | |
4UUG
 
 | | The (R)-selective amine transaminase from Aspergillus fumigatus with inhibitor bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Thomsen, M, Hinrichs, W. | | Deposit date: | 2014-07-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Dual Substrate Recognition of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus
FEBS J., 282, 2015
|
|
7T7I
 
 | | EBV nuclear egress complex | | Descriptor: | Nuclear egress protein 1, Nuclear egress protein 2, ZINC ION | | Authors: | Thorsen, M.K, Heldwein, E.E. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | The nuclear egress complex of Epstein-Barr virus buds membranes through an oligomerization-driven mechanism.
Plos Pathog., 18, 2022
|
|
7SMP
 
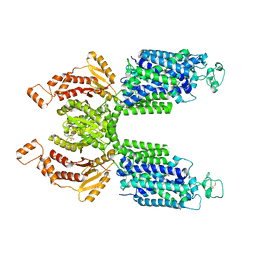 | | CryoEM structure of NKCC1 Bu-I | | Descriptor: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
8RP8
 
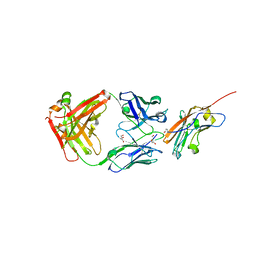 | | Structure of K2 Fab in complex with human CD47 ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Laursen, M, Kelpsas, V, Rose, N. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1.
Mabs, 16, 2024
|
|
7NP8
 
 | |
8STE
 
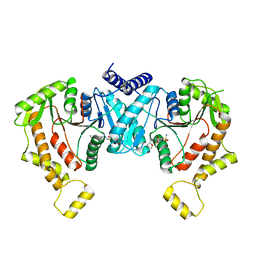 | | Cryo-EM structure of NKCC1 Fu_CTD | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
5L3A
 
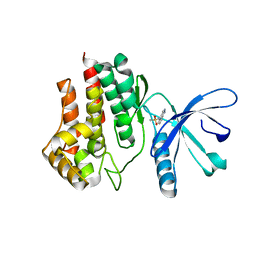 | | Fragment-based discovery of 6-arylindazole JAK inhibitors | | Descriptor: | Tyrosine-protein kinase JAK2, ~{N}-(1~{H}-indazol-4-yl)methanesulfonamide | | Authors: | Soerensen, M.D, Dack, K.N, Greve, D.R, Ritzen, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Fragment-Based Discovery of 6-Arylindazole JAK Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
4WCU
 
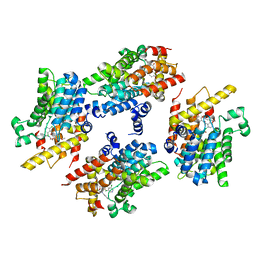 | | PDE4 complexed with inhibitor | | Descriptor: | MAGNESIUM ION, N-benzyl-2-{6-[(3,5-dichloropyridin-4-yl)acetyl]-2,3-dimethoxyphenoxy}acetamide, ZINC ION, ... | | Authors: | Sorensen, M.D. | | Deposit date: | 2014-09-05 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Early Clinical Development of 2-{6-[2-(3,5-Dichloro-4-pyridyl)acetyl]-2,3-dimethoxyphenoxy}-N-propylacetamide (LEO 29102), a Soft-Drug Inhibitor of Phosphodiesterase 4 for Topical Treatment of Atopic Dermatitis.
J. Med. Chem., 57, 2014
|
|
4W1O
 
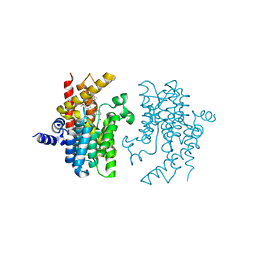 | | PDE4D complexed with inhibitor | | Descriptor: | N-(3,5-dichloropyridin-4-yl)-3-[(3-ethyl-1,2-oxazol-5-yl)methoxy]-4-methoxybenzamide, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Sorensen, M.D. | | Deposit date: | 2014-08-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Early Clinical Development of 2-{6-[2-(3,5-Dichloro-4-pyridyl)acetyl]-2,3-dimethoxyphenoxy}-N-propylacetamide (LEO 29102), a Soft-Drug Inhibitor of Phosphodiesterase 4 for Topical Treatment of Atopic Dermatitis
J.Med.Chem, 57, 2014
|
|
7C5V
 
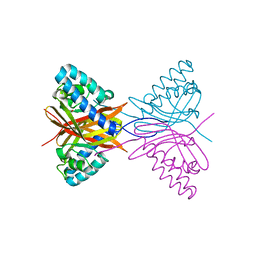 | |
7C5W
 
 | |
7C5Y
 
 | |
7C5X
 
 | |
7DNN
 
 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNM
 
 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DVE
 
 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EM8
 
 | | Crystal structure of the PI5P4Kbeta T201M-2a-ATP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM5
 
 | | Crystal structure of the PI5P4Kbeta F205L-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
