2PUE
 
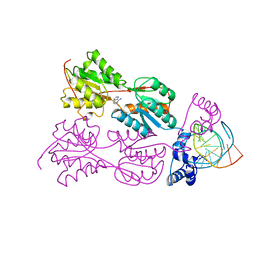 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | ADENINE, DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), PROTEIN (PURINE REPRESSOR ) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
4OVW
 
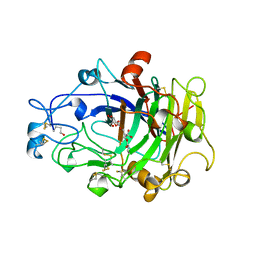 | | ENDOGLUCANASE I COMPLEXED WITH EPOXYBUTYL CELLOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(beta-D-glucopyranosyloxy)-2,2-dihydroxybutyl propanoate, ENDOGLUCANASE I | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
3GXQ
 
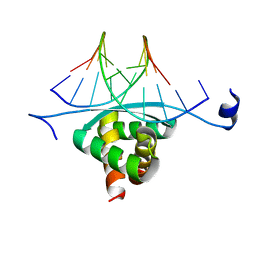 | | Structure of ArtA and DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*AP*CP*AP*TP*G)-3'), DNA (5'-D(*AP*CP*AP*TP*GP*TP*CP*AP*TP*GP*T)-3'), Putative regulator of transfer genes ArtA | | Authors: | Ni, L, Firth, N, Schumacher, M.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes.
Nucleic Acids Res., 37, 2009
|
|
1OJJ
 
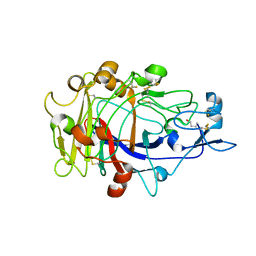 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
3NXC
 
 | |
2VE4
 
 | | Substrate free cyanobacterial CYP120A1 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 120 | | Authors: | Kuhnel, K, Ke, N, Sligar, S.G, Schuler, M.A, Schlichting, I. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Substrate-Free and Retinoic Acid-Bound Cyanobacterial Cytochrome P450 Cyp120A1.
Biochemistry, 47, 2008
|
|
2MF0
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
3M8E
 
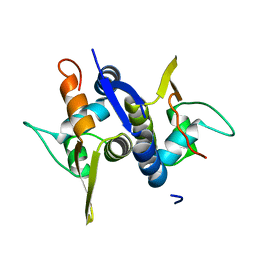 | | Protein structure of Type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Ni, L, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HTA
 
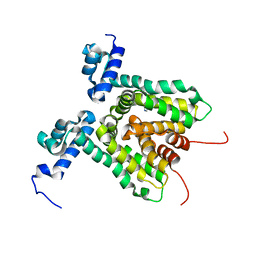 | |
3HTI
 
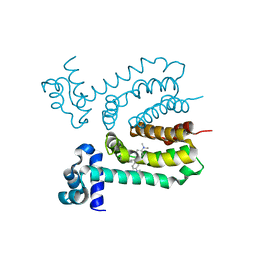 | |
4CE5
 
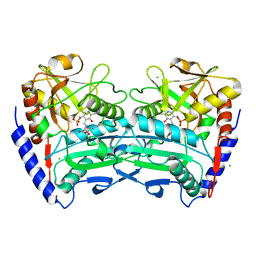 | | First crystal structure of an (R)-selective omega-transaminase from Aspergillus terreus | | Descriptor: | AT-OMEGATA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lyskowski, A, Gruber, C, Steinkellner, G, Schurmann, M, Schwab, H, Gruber, K, Steiner, K. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of an (R)-Selective Omega-Transaminase from Aspergillus Terreus
Plos One, 9, 2014
|
|
2ENG
 
 | | ENDOGLUCANASE V | | Descriptor: | ENDOGLUCANASE V | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1995-06-29 | | Release date: | 1996-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 A resolution.
Biochemistry, 34, 1995
|
|
6HLL
 
 | | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist CP-99,994 | | Descriptor: | (2~{S},3~{S})-~{N}-[(2-methoxyphenyl)methyl]-2-phenyl-piperidin-3-amine, Substance-P receptor,GlgA glycogen synthase,Substance-P receptor | | Authors: | Schoppe, J, Ehrenmann, J, Klenk, C, Rucktooa, P, Schutz, M, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists.
Nat Commun, 10, 2019
|
|
1BDI
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Glasfeld, A, Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-07-25 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Positively Charged Residue Bound in the Minor Groove Does not Alter the Bending of a DNA Duplex
J.Am.Chem.Soc., 118, 1996
|
|
1PRV
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1PRU
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
2BVW
 
 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1999-02-18 | | Release date: | 2000-02-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding.
Biochemistry, 38, 1999
|
|
1OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH NON-HYDROLYSABLE SUBSTRATE ANALOGUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1,4-dithio-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group.
Biochemistry, 35, 1996
|
|
3OVW
 
 | | ENDOGLUCANASE I NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
3M89
 
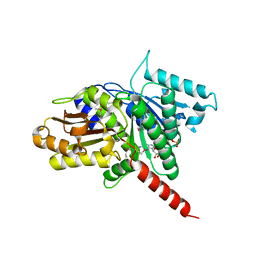 | | Structure of TubZ-GTP-g-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, FtsZ/tubulin-related protein | | Authors: | Ni, L, Xu, W, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PUG
 
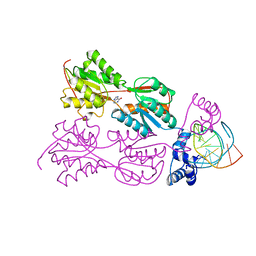 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
2PUF
 
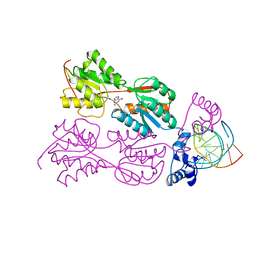 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
1BDH
 
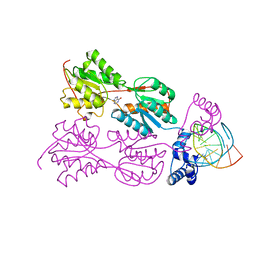 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Glasfeld, A, Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-07-25 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Residue Bound in the Minor Groove Does not Alter the Bending of a DNA Duplex
J.Am.Chem.Soc., 118, 1996
|
|
1QH6
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
