4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCO
 
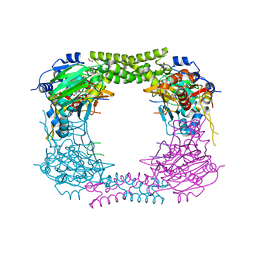 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 S373C) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G214C) peptide fragment (complex-3) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCM
 
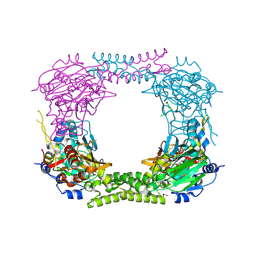 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-1) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4BXF
 
 | |
2BRT
 
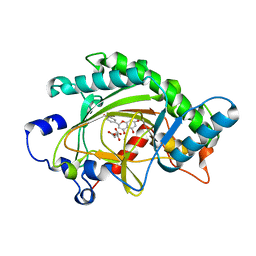 | | ANTHOCYANIDIN SYNTHASE FROM ARABIDOPSIS THALIANA COMPLEXED with naringenin | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, LEUCOANTHOCYANIDIN DIOXYGENASE, ... | | Authors: | Turnbull, J.J, Clifton, I.J, Welford, R.W.D, Schofield, C.J. | | Deposit date: | 2005-05-11 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Studies on Anthocyanidin Synthase Catalysed Oxidation of Flavanone Substrates: The Effect of C-2 Stereochemistry on Product Selectivity and Mechanism
Org.Biomol.Chem., 3, 2005
|
|
2CGO
 
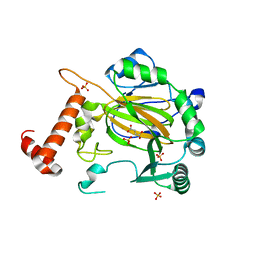 | | FACTOR INHIBITING HIF-1 ALPHA with fumarate | | Descriptor: | FE (III) ION, FUMARIC ACID, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA INHIBITOR, ... | | Authors: | McDonough, M.A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2006-03-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies on the Inhibition of the Hypoxia-Inducible Transcription Factor Hydroxylases by Tricarboxylic Acid Cycle Intermediates.
J.Biol.Chem., 282, 2007
|
|
2CGN
 
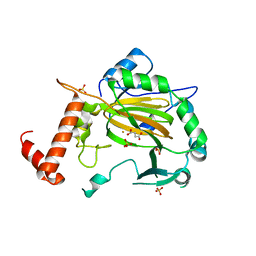 | | FACTOR INHIBITING HIF-1 ALPHA with succinate | | Descriptor: | FE (III) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA INHIBITOR, SUCCINIC ACID, ... | | Authors: | McDonough, M.A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2006-03-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Studies on the Inhibition of the Hypoxia-Inducible Transcription Factor Hydroxylases by Tricarboxylic Acid Cycle Intermediates.
J.Biol.Chem., 282, 2007
|
|
2XUM
 
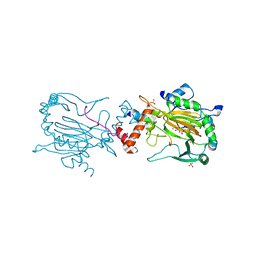 | | FACTOR INHIBITING HIF (FIH) Q239H MUTANT IN COMPLEX WITH ZN(II), NOG AND ASP-SUBSTRATE PEPTIDE (20-MER) | | Descriptor: | ASP-SUBSTRATE PEPTIDE 2, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor.
J. Biol. Chem., 286, 2011
|
|
4IE5
 
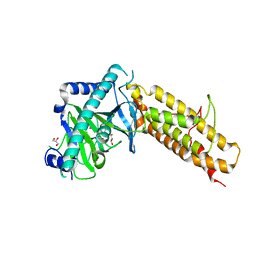 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with N-[(3-hydroxypyridin-2-yl)carbonyl]glycine (MD6) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, N-[(3-hydroxypyridin-2-yl)carbonyl]glycine, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
4IE7
 
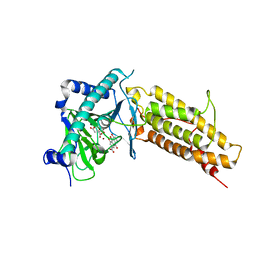 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with citrate and rhein (RHN) | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, CITRATE ANION, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6004 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
4IE6
 
 | |
4IDZ
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with N-oxalylglycine (NOG) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
6RBJ
 
 | | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, CHLORIDE ION, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
4IE4
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with 5-carboxy-8-hydroxyquinoline (IOX1) | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5048 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
6RZ0
 
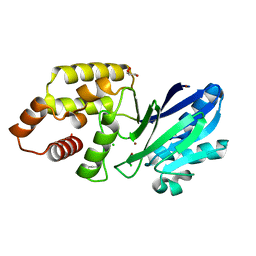 | |
6RMF
 
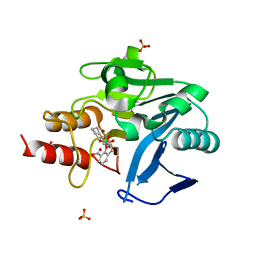 | | Crystal structure of NDM-1 with VNRX-5133 | | Descriptor: | (10aR)-2-(((1r,4R)-4-((2-aminoethyl)amino)cyclohexyl)methyl)-6-carboxy-4-hydroxy-4,10a-dihydro-10H-benzo[5,6][1,2]oxaborinino[2,3-b][1,4,2]oxazaborol-4-uide, (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J, Schofield, C.J. | | Deposit date: | 2019-05-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Bicyclic Boronate VNRX-5133 Inhibits Metallo- and Serine-beta-Lactamases.
J.Med.Chem., 62, 2019
|
|
6RPN
 
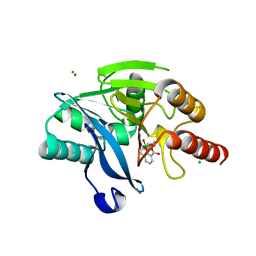 | | Structure of metallo beta lactamase VIM-2 with cyclic boronate APC308. | | Descriptor: | (3~{R})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, (3~{S})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, Beta-lactamase VIM-2, ... | | Authors: | Parkova, A, Lucic, A, Brem, J, McDonough, M.A, Langley, G.W, Schofield, C.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Broad Spectrum beta-Lactamase Inhibition by a Thioether Substituted Bicyclic Boronate.
Acs Infect Dis., 6, 2020
|
|
4IW3
 
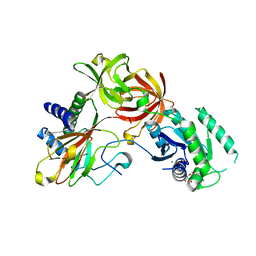 | | Crystal structure of a Pseudomonas putida prolyl-4-hydroxylase (P4H) in complex with elongation factor Tu (EF-Tu) | | Descriptor: | Elongation factor Tu-A, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4J0Q
 
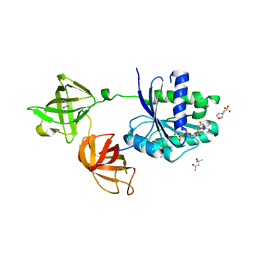 | | Crystal structure of Pseudomonas putida elongation factor Tu (EF-Tu) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Elongation factor Tu-A, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4J25
 
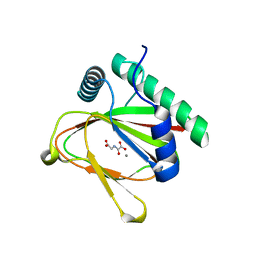 | |
6RUJ
 
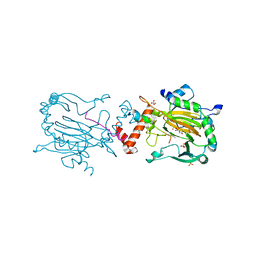 | |
4JAA
 
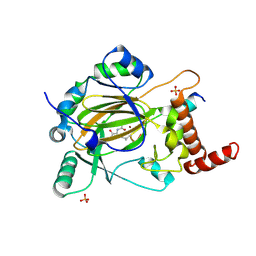 | | Factor inhibiting HIF-1 alpha in complex with consensus ankyrin repeat domain-(d)LEU peptide | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-(d)LEU, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Scotti, J.S, Ge, W, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an oxygenase in complex with substrate
To be Published
|
|
6T5Y
 
 | | Crystal structure of AmpC from E.coli with Zidebactam (WCK 5107) | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
6T3D
 
 | | Crystal structure of AmpC from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
4JHT
 
 | | Crystal Structure of AlkB in complex with 5-carboxy-8-hydroxyquinoline (IOX1) | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, ACETATE ION, Alpha-ketoglutarate-dependent dioxygenase AlkB, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation.
Chem Sci, 4, 2013
|
|
