3QZE
 
 | |
3PBI
 
 | |
3PBC
 
 | |
2Q3B
 
 | |
5OLT
 
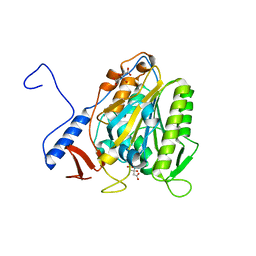 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Vella, P, Polyakova, A, Lindqvist, Y, Schnell, R, Bourenkov, G, Schneider, T, Schneider, G. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
5IW8
 
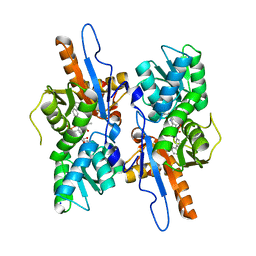 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 4 [5-(3-([1,1'-Biphenyl]-3-yl)ureido)-2-hydroxybenzoic acid] | | Descriptor: | 5-{[([1,1'-biphenyl]-3-yl)carbamoyl]amino}-2-hydroxybenzoic acid, O-phosphoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Brunner, K, Schnell, R, Schneider, G. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
4AG3
 
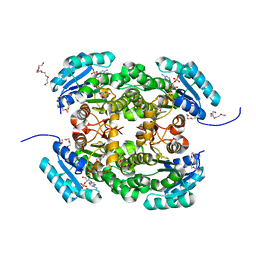 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with NADPH at 1.8A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | Authors: | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | Deposit date: | 2012-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
5OJH
 
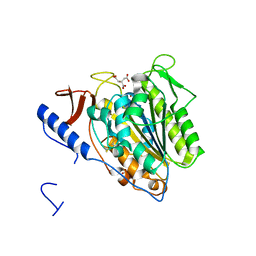 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRATE ANION, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Schneider, G, Vella, P, Lindqvist, Y, Schnell, R. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
4RFV
 
 | | Structure of the Mycobacterium tuberculosis APS kinase CysC Cys556Ala mutant | | Descriptor: | Bifunctional enzyme CysN/CysC, PHOSPHATE ION | | Authors: | Poyraz, O, Brunner, K, Schnell, R, Schneider, G. | | Deposit date: | 2014-09-28 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structures of the Kinase Domain of the Sulfate-Activating Complex in Mycobacterium tuberculosis.
Plos One, 10, 2015
|
|
4UMJ
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound ibandronic acid molecules. | | Descriptor: | GERANYLTRANSTRANSFERASE, IBANDRONATE, MAGNESIUM ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2014-05-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZEI
 
 | | Structure of the Mycobacterium tuberculosis O-Acetylserine Sulfhydrylase (OASS) CysK1 in complex with a small molecule inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(Z)-[(5Z)-5-[[2-(2-hydroxy-2-oxoethyloxy)phenyl]methylidene]-3-methyl-4-oxidanylidene-1,3-thiazolidin-2-ylidene]amino]benzoic acid, O-ACETYLSERINE SULFHYDRYLASE, ... | | Authors: | Poyraz, O, Schnell, R, Schneider, G. | | Deposit date: | 2012-12-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Novel Thiazolidine Inhibitors of O-Acetyl Serine Sulfhydrylase from Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
3ZCD
 
 | |
3ZMB
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696. | | Descriptor: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZOU
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696, and substrate geranyl pyrophosphate. | | Descriptor: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, DIMETHYL SULFOXIDE, FARNESYL PYROPHOSPHATE SYNTHASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZL6
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PAO1, with bound fragment KM10833. | | Descriptor: | 2-(1,2-benzoxazol-3-yl)ethanoic acid, DIMETHYL SULFOXIDE, GERANYLTRANSTRANSFERASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-01-28 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZMC
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound substrate molecule Geranyl pyrophosphate. | | Descriptor: | DIMETHYL SULFOXIDE, GERANYL DIPHOSPHATE, GERANYLTRANSTRANSFERASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5LB1
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with thionitrobenzoate (TNB) adduct at the active site cysteine-354 | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
5LBG
 
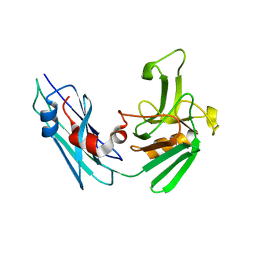 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with faropenem-derived adduct at the active site cysteine-354 | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
5EZ7
 
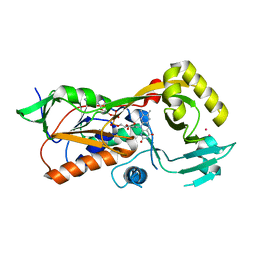 | | Crystal structure of the FAD dependent oxidoreductase PA4991 from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MERCURY (II) ION, flavoenzyme PA4991 | | Authors: | Jacewicz, A, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the flavoenzyme PA4991 from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3ZQU
 
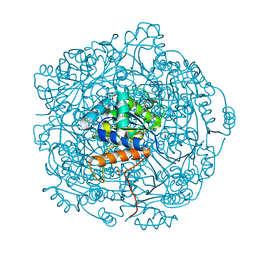 | | STRUCTURE OF A PROBABLE AROMATIC ACID DECARBOXYLASE | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, PROBABLE AROMATIC ACID DECARBOXYLASE, SULFATE ION | | Authors: | Kopec, J, Schnell, R, Schneider, G. | | Deposit date: | 2011-06-11 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Pa4019, a Putative Aromatic Acid Decarboxylase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3DKI
 
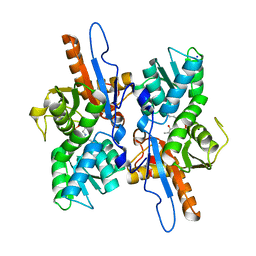 | |
3FGP
 
 | |
3EUL
 
 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4LJ1
 
 | | RipD (Rv1566c) from Mycobacterium tuberculosis: a non-catalytic NlpC/p60 domain protein with two penta-peptide repeat units (PVQQA-PVQPA) | | Descriptor: | Invasion-associated protein | | Authors: | Both, D, Steiner, E.M, Linder, D.C, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-04 | | Release date: | 2013-11-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | RipD (Rv1566c) from Mycobacterium tuberculosis: adaptation of an NlpC/p60 domain to a non-catalytic peptidoglycan-binding function.
Biochem.J., 457, 2014
|
|
2Q3D
 
 | |
