7MM8
 
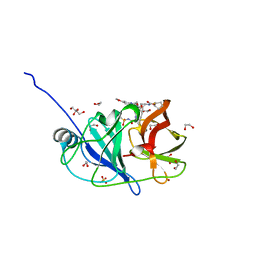 | | Crystal structure of HCV NS3/4A protease in complex with NR02-08 | | 分子名称: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6C2O
 
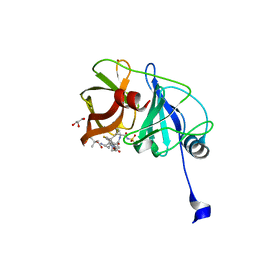 | | Crystal structure of HCV NS3/4A protease variant Y56H in complex with danoprevir | | 分子名称: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, GLYCEROL, NS3 protease, ... | | 著者 | Matthew, A.N, Schiffer, C.A. | | 登録日 | 2018-01-08 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.179 Å) | | 主引用文献 | Clinical signature variant of HCV NS3/4A protease uses a novel mechanism to confer resistance
To be Published
|
|
6W6R
 
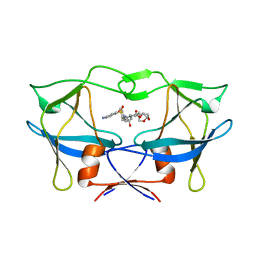 | | WT HTLV-1 Protease in Complex with UMass6 (UM6) | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, HTLV-1 Protease | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
6W6S
 
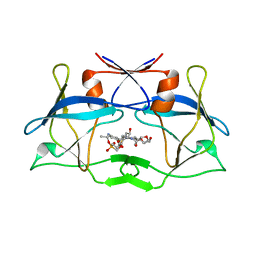 | | WT HTLV-1 Protease in Complex with Phosphonated UMass6 (PU6) | | 分子名称: | HTLV-1 Protease, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
6C2N
 
 | | Crystal structure of HCV NS3/4A double mutant variant Y56H/D168A in complex with danoprevir | | 分子名称: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS4A COFACTOR-NS3 PROTEIN CHIMERA, SULFATE ION, ... | | 著者 | Matthew, A.N, Schiffer, C.A. | | 登録日 | 2018-01-08 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Clinical signature variant of HCV NS3/4A protease uses a novel mechanism to confer resistance
To Be Published
|
|
6UE3
 
 | |
2FGV
 
 | | X-ray crystal structure of HIV-1 Protease T80N variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | 分子名称: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | 著者 | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | 登録日 | 2005-12-22 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
2FGU
 
 | | X-ray crystal structure of HIV-1 Protease T80S variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | 分子名称: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | 著者 | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | 登録日 | 2005-12-22 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
2F3K
 
 | | Substrate envelope and drug resistance: crystal structure of r01 in complex with wild-type hiv-1 protease | | 分子名称: | (3S,4AS,8AS)-N-(TERT-BUTYL)-2-[(3S)-3-({3-(METHYLSULFONYL)-N-[(PYRIDIN-3-YLOXY)ACETYL]-L-VALYL}AMINO)-2-OXO-4-PHENYLBUTYL]DECAHYDROISOQUINOLINE-3-CARBOXAMIDE, PHOSPHATE ION, Protease | | 著者 | Prabu-Jeyabalan, M, King, N.M, Nalivaika, E.A, Heilek-Snyder, G, Cammack, N, Schiffer, C.A. | | 登録日 | 2005-11-21 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease.
Antimicrob.Agents Chemother., 50, 2006
|
|
6VDN
 
 | |
6VDL
 
 | | HCV NS3/4A protease A156T mutant in complex with glecaprevir | | 分子名称: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, GLYCEROL, ... | | 著者 | Timm, J, Schiffer, C.A. | | 登録日 | 2019-12-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | RAS at position 156 of HCV NS3/4A protease abolish inhibition by current HCV drugs
To Be Published
|
|
6VDM
 
 | | HCV NS3/4A protease A156T, D168E double mutant in complex with glecaprevir | | 分子名称: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, GLYCEROL, ... | | 著者 | Timm, J, Schiffer, C.A. | | 登録日 | 2019-12-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | RAS at position 156 of HCV NS3/4A protease abolish inhibition by current HCV drugs
To Be Published
|
|
6VDO
 
 | |
4Q1Y
 
 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | 著者 | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | 登録日 | 2014-04-04 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
4QJ8
 
 | |
4IOU
 
 | |
4QJ6
 
 | |
4QJ9
 
 | |
6WF1
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H10 Hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | 著者 | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | 登録日 | 2020-04-03 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.19 Å) | | 主引用文献 | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6DH5
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass6 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DGZ
 
 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass6 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH0
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.899 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH8
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass6 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
4QJ7
 
 | |
6W6Q
 
 | | WT HTLV-1 Protease in Complex with Darunavir (DRV) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HTLV-1 Protease | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2020-03-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | To Be Determined
To Be Published
|
|
