8Q2N
 
 | |
5OS9
 
 | |
4RZL
 
 | |
8EXA
 
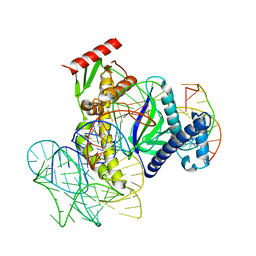 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8EX9
 
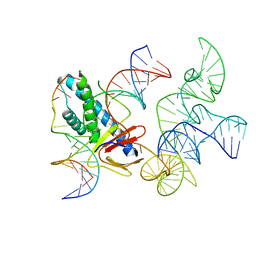 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8BF8
 
 | | ISDra2 TnpB in complex with reRNA | | Descriptor: | Deinococcus radiodurans R1 chromosome 1, RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Siksnys, V, Montoya, G, Druteika, G, Silanskas, A, Venclovas, C, Karvelis, T, Kazlauskas, D. | | Deposit date: | 2022-10-24 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8PJ9
 
 | |
8PK1
 
 | | Cas1-Cas2 CRISPR integrase bound to prespacer DNA, Streptococcus thermophilus DGCC 7710 CRISPR3 system | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, Chains: G,J, ... | | Authors: | Sasnauskas, G, Gaizauskaite, U, Tamulaitiene, G. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for spacer acquisition in a type II-A CRISPR-Cas system
To Be Published
|
|
8PM4
 
 | | Cryo-EM structure of the Cas12m-crRNA-target DNA complex | | Descriptor: | DNA oligoduplex, non-target strand, chain D, ... | | Authors: | Sasnauskas, G, Tamulaitiene, G, Karvelis, T, Bigelyte, G, Siksnys, V. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing.
Nucleic Acids Res., 52, 2024
|
|
6FAS
 
 | | Crystal structure of VAL1 B3 domain in complex with cognate DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*GP*CP*CP*AP*TP*GP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*GP*CP*AP*TP*GP*GP*CP*T)-3') | | Authors: | Sasnauskas, G. | | Deposit date: | 2017-12-17 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1.
Nucleic Acids Res., 46, 2018
|
|
6GCF
 
 | |
6GCE
 
 | |
6GCD
 
 | |
1KNV
 
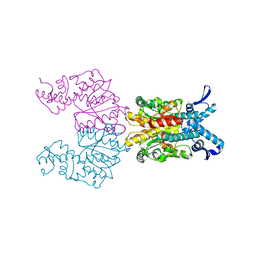 | | Bse634I restriction endonuclease | | Descriptor: | ACETATE ION, Bse634I restriction endonuclease, CHLORIDE ION | | Authors: | Grazulis, S, Deibert, M, Rimseliene, R, Skirgaila, R, Sasnauskas, G, Lagunavicius, A, Repin, V, Urbanke, C, Huber, R, Siksnys, V. | | Deposit date: | 2001-12-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence.
Nucleic Acids Res., 30, 2002
|
|
8BTP
 
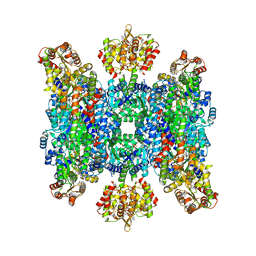 | | Helical structure of BcThsA in complex with 1''-3'gc(etheno)ADPR | | Descriptor: | (1S,3S,4R,5R,7R,15R,16S,17R)-5-imidazo[2,1-f]purin-3-yl-10,12-bis(oxidanyl)-10,12-bis(oxidanylidene)-2,6,9,11,13,18-hexaoxa-10$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{3,7}]octadecane-4,16,17-triol, ETHENO-NAD, NAD(+) hydrolase ThsA | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
5O63
 
 | |
3ZI5
 
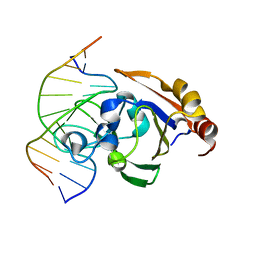 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | Descriptor: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | Authors: | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2013-01-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
1FIU
 
 | | TETRAMERIC RESTRICTION ENDONUCLEASE NGOMIV IN COMPLEX WITH CLEAVED DNA | | Descriptor: | ACETIC ACID, DNA (5'-D(*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*CP*GP*C)-3'), ... | | Authors: | Deibert, M, Grazulis, S, Sasnauskas, G, Siksnys, V, Huber, R. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Nat.Struct.Biol., 7, 2000
|
|
9EYJ
 
 | |
9EYI
 
 | |
9EYK
 
 | |
8BTO
 
 | | Helical structure of BcThsA in complex with 1''-3'gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, NAD(+) hydrolase ThsA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
6T22
 
 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5hmCGA target sequence | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*(5HC)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5HC)P*GP*AP*TP*TP*C)-3'), EcoKMcrA modification dependent restriction endonuclease | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-10-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
6T21
 
 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5mCGA target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*TP*(5CM)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5CM)P*GP*AP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-10-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
6R64
 
 | | N-terminal domain of modification dependent EcoKMcrA restriction endonuclease (NEco) in complex with C5mCGG target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*(5CM)P*GP*GP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
