2A25
 
 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
2A26
 
 | | Crystal structure of the N-terminal, dimerization domain of Siah Interacting Protein | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Calcyclin-binding protein, SULFATE ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
6UZT
 
 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
1EGW
 
 | | CRYSTAL STRUCTURE OF MEF2A CORE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, ... | | Authors: | Santelli, E, Richmond, T.J. | | Deposit date: | 2000-02-17 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of MEF2A core bound to DNA at 1.5 A resolution.
J.Mol.Biol., 297, 2000
|
|
1T6B
 
 | | Crystal structure of B. anthracis Protective Antigen complexed with human Anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Santelli, E, Bankston, L.A, Leppla, S.H, Liddington, R.C. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a complex between anthrax toxin and its host cell receptor
Nature, 430, 2004
|
|
4I7D
 
 | | Siah1 bound to synthetic peptide (ACE)KLRPVAMVRP(PRK)VR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
2O3O
 
 | |
4I7B
 
 | | Siah1 bound to synthetic peptide (ACE)KLRPV(ABA)MVRPTVR | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ZINC ION | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I7C
 
 | | Siah1 mutant bound to synthetic peptide (ACE)KLRPV(23P)MVRPWVR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
8FBZ
 
 | | Crystal Structure of apo human Glutathione Synthetase Y270E | | Descriptor: | GLYCEROL, Glutathione synthetase, SULFATE ION | | Authors: | Stanford, S.M, Santelli, E, Sankaran, B, Murali, R, Bottini, N. | | Deposit date: | 2022-11-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting prostate tumor low-molecular weight tyrosine phosphatase for oxidation-sensitizing therapy.
Sci Adv, 10, 2024
|
|
2QPF
 
 | | Crystal Structure of Mouse Transthyretin | | Descriptor: | Transthyretin | | Authors: | Reixach, N, Foss, T.R, Santelli, E, Pascual, J, Kelly, J.W, Buxbaum, J.N. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human-Murine Transthyretin Heterotetramers Are Kinetically Stable and Non-amyloidogenic: A LESSON IN THE GENERATION OF TRANSGENIC MODELS OF DISEASES INVOLVING OLIGOMERIC PROTEINS.
J.Biol.Chem., 283, 2008
|
|
1Y19
 
 | | Structural basis for phosphatidylinositol phosphate kinase type I-gamma binding to talin at focal adhesions | | Descriptor: | Phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma, Talin 1 | | Authors: | de Pereda, J.M, Wegener, K, Santelli, E, Bate, N, Ginsberg, M.H, Critchley, D.R, Campbell, I.D, Liddington, R.C. | | Deposit date: | 2004-11-17 | | Release date: | 2005-01-04 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural bases for phosphatidylinositol phosphate kinase type I-gamma binding to talin at focal adhesions
J.Biol.Chem., 280, 2005
|
|
3FKU
 
 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Neutralizing antibody F10, ... | | Authors: | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
7KH8
 
 | | Human LMPTP in complex with inhibitor | | Descriptor: | 3-[(2,6-dichlorophenyl)methyl]-8-(2-methylphenyl)-3H-purin-6-amine, Low molecular weight phosphotyrosine protein phosphatase, NITRATE ION | | Authors: | Stanford, S.M, Diaz, M.A, Ardecky, R.J, Zou, J, Roosild, T, Holmes, Z.J, Hedrick, M.P, Rodiles, S, Santelli, E, Chung, T.D.Y, Jackson, M.R, Bottini, N, Pinkerton, A.B. | | Deposit date: | 2020-10-20 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Orally Bioavailable Purine-Based Inhibitors of the Low-Molecular-Weight Protein Tyrosine Phosphatase.
J.Med.Chem., 64, 2021
|
|
3H16
 
 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
3PMC
 
 | | Crystal structure of the sporulation inhibitor pXO2-61 from Bacillus anthracis | | Descriptor: | CHLORIDE ION, IODIDE ION, Uncharacterized protein pXO2-61/BXB0075/GBAA_pXO2_0075 | | Authors: | Stranzl, G.R, Santelli, E, Bankston, L.A, La Clair, C, Bobkov, A, Schwarzenbacher, R, Godzik, A, Perego, M, Grynberg, M, Liddington, R.C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Insights into Inhibition of Bacillus anthracis Sporulation by a Novel Class of Non-heme Globin Sensor Domains.
J.Biol.Chem., 286, 2011
|
|
3JRN
 
 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
2GHW
 
 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | Descriptor: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2GHV
 
 | | Crystal structure of SARS spike protein receptor binding domain | | Descriptor: | Spike glycoprotein | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2OTN
 
 | | Crystal structure of the catalytically active form of diaminopimelate epimerase from Bacillus anthracis | | Descriptor: | Diaminopimelate epimerase | | Authors: | Matho, M.H, Fukuda, K, Santelli, E, Jaroszewski, L, Liddington, R.C, Roper, D. | | Deposit date: | 2007-02-08 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and inhibition of a catalytically active form of diaminopimelate epimerase (DapF)from Bacillus anthracis
To be Published
|
|
3PMD
 
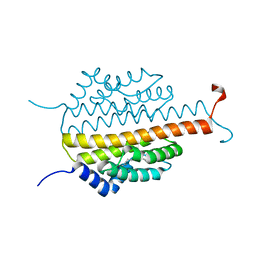 | | Crystal structure of the sporulation inhibitor pXO1-118 from Bacillus anthracis | | Descriptor: | CHLORIDE ION, Conserved domain protein, UNDECANOIC ACID | | Authors: | Stranzl, G.R, Santelli, E, Bankston, L.A, La Clair, C, Bobkov, A, Schwarzenbacher, R, Godzik, A, Perego, M, Grynberg, M, Liddington, R.C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Inhibition of Bacillus anthracis Sporulation by a Novel Class of Non-heme Globin Sensor Domains.
J.Biol.Chem., 286, 2011
|
|
