7S71
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI35 | | 分子名称: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6W
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | 分子名称: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S73
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI37 | | 分子名称: | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name), 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | 分子名称: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6X
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI30 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S72
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
4F6H
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | 分子名称: | Beta-lactamase, PHOSPHATE ION, SULFATE ION, ... | | 著者 | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2012-05-14 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.744 Å) | | 主引用文献 | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
4EZ1
 
 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with alpha-conotoxin BuIA | | 分子名称: | Alpha-conotoxin BuIA, MANGANESE (II) ION, Soluble acetylcholine receptor | | 著者 | Talley, T.T, Reger, A.S, Kim, C, Sankaran, B, Ho, K, Taylor, P, McIntosh, J.M. | | 登録日 | 2012-05-02 | | 公開日 | 2013-05-08 | | 最終更新日 | 2013-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Pairwise interaction of alpha-conotoxin BuIA Pro6 with the beta subunit of nicotinic acetylcholine receptor
To be Published
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | 分子名称: | Beta-lactamase, CITRATE ANION, ZINC ION | | 著者 | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2012-05-15 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
7K2W
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed cefotaxime | | 分子名称: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | 著者 | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | 登録日 | 2020-09-09 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7K2Y
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed ampicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase | | 著者 | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | 登録日 | 2020-09-09 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7KHY
 
 | | Crystal structure of OXA-163 K73A in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-22 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KH9
 
 | | Crystal structure of OXA-48 K73A in complex with imipenem | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase, CHLORIDE ION | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-20 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KHZ
 
 | | Crystal structure of OXA-163 K73A in complex with imipenem | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-22 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KHQ
 
 | | Crystal structure of OXA-48 K73A in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-21 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7K2X
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase | | 分子名称: | Beta-lactamase, GLYCEROL | | 著者 | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | 登録日 | 2020-09-09 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
3LT0
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T1 | | 分子名称: | 4-(2,4-dichlorophenoxy)-3-hydroxybenzaldehyde, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Maity, K, Bhargav, S.P, Sankaran, B, Surolia, N, Surolia, A, Suguna, K. | | 登録日 | 2010-02-14 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3LT1
 
 | | Enoyl-ACP Reductase from Plasmodium falciparum (PfENR) in complex with triclosan variant T2 | | 分子名称: | 5-(chloromethyl)-2-(2,4-dichlorophenoxy)phenol, Enoyl-ACP reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Maity, K, Bhargav, S.P, Sankaran, B, Surolia, N, Surolia, A, Suguna, K. | | 登録日 | 2010-02-14 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray crystallographic analysis of the complexes of enoyl acyl carrier protein reductase of Plasmodium falciparum with triclosan variants to elucidate the importance of different functional groups in enzyme inhibition
Iubmb Life, 62, 2010
|
|
3NMD
 
 | | Crystal structure of the leucine zipper domain of cGMP dependent protein kinase I beta | | 分子名称: | GLYCEROL, HEXANE-1,6-DIOL, cGMP Dependent PRotein Kinase | | 著者 | Kim, C, Casteel, D.E, Smith-Nguyen, E.V, Sankaran, B, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2010-06-22 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | A crystal structure of the cyclic GMP-dependent protein kinase I{beta} dimerization/docking domain reveals molecular details of isoform-specific anchoring.
J.Biol.Chem., 285, 2010
|
|
3OF1
 
 | | Crystal Structure of Bcy1, the Yeast Regulatory Subunit of PKA | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase regulatory subunit | | 著者 | Rinaldi, J, Wu, J, Yang, J, Ralston, C.Y, Sankaran, B, Moreno, S, Taylor, S.S. | | 登録日 | 2010-08-13 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of Yeast Regulatory Subunit: A Glimpse into the Evolution of PKA Signaling.
Structure, 18, 2010
|
|
3Q6L
 
 | | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution | | 分子名称: | Fatty acid-binding protein, adipocyte | | 著者 | Prasad, G.S, Carney, D, Small, V, Sankaran, B, Zwart, P.H. | | 登録日 | 2011-01-02 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution
To be Published
|
|
3QHY
 
 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | 分子名称: | Beta-lactamase, Beta-lactamase inhibitory protein II | | 著者 | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2011-01-26 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3SEJ
 
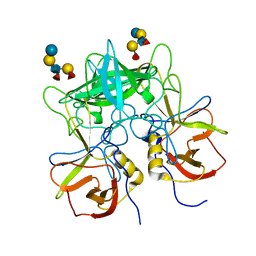 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to Secretor Lewis HBGA (LeB) | | 分子名称: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | 登録日 | 2011-06-10 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.041 Å) | | 主引用文献 | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SLD
 
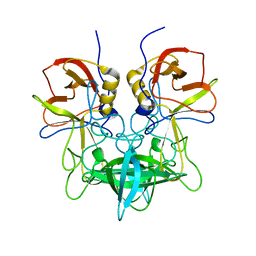 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | 分子名称: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | 著者 | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | 登録日 | 2011-06-24 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.679 Å) | | 主引用文献 | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SI9
 
 | | Crystal structure of Dihydrodipicolinate Synthase from Bartonella Henselae | | 分子名称: | 1,2-ETHANEDIOL, Dihydrodipicolinate synthase | | 著者 | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Abendroth, J, Sankaran, B. | | 登録日 | 2011-06-17 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cloning, expression, purification, crystallization and X-ray diffraction analysis of dihydrodipicolinate synthase from the human pathogenic bacterium Bartonella henselae strain Houston-1 at 2.1 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
