2F2E
 
 | | Crystal Structure of PA1607, a Putative Transcription Factor | | Descriptor: | PA1607, SULFATE ION, alpha-D-glucopyranose | | Authors: | Sieminska, E.A, Xu, X, Zheng, H, Lunin, V, Cuff, M, Joachimiak, A, Edwards, A, Savchenko, A, Sanders, D.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-16 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The X-ray crystal structure of PA1607 from Pseudomonas aureginosa at 1.9 A resolution--a putative transcription factor.
Protein Sci., 16, 2007
|
|
2IIR
 
 | |
3MJ4
 
 | | Crystal structure of UDP-galactopyranose mutase in complex with phosphonate analog of UDP-galactopyranose | | Descriptor: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Karunan Partha, S, Sadeghi-Khomami, A, Slowski, K, Kotake, T, Thomas, N.R, Jakeman, D.L, Sanders, D.A.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemoenzymatic Synthesis, Inhibition Studies, and X-ray Crystallographic Analysis of the Phosphono Analog of UDP-Galp as an Inhibitor and Mechanistic Probe for UDP-Galactopyranose Mutase.
J.Mol.Biol., 403, 2010
|
|
3MZ0
 
 | | Crystal structure of apo myo-inositol dehydrogenase from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-05-11 | | Release date: | 2010-09-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3HDQ
 
 | |
3HE3
 
 | |
3HDY
 
 | | Crystal Structure of UDP-galactopyranose mutase (reduced form) in complex with substrate | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Partha, S.K, van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate binding to UDP-galactopyranose mutase: crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP.
J.Mol.Biol., 394, 2009
|
|
5F1V
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
5F1U
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
6MOS
 
 | |
1G99
 
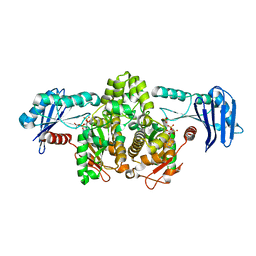 | | AN ANCIENT ENZYME: ACETATE KINASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | ACETATE KINASE, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Buss, K.A, Cooper, D.R, Ingram-Smith, C, Ferry, J.G, Sanders, D.A, Hasson, M.S. | | Deposit date: | 2000-11-22 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases.
J.Bacteriol., 183, 2001
|
|
4K2M
 
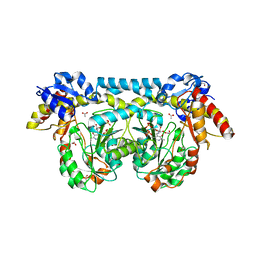 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4K2B
 
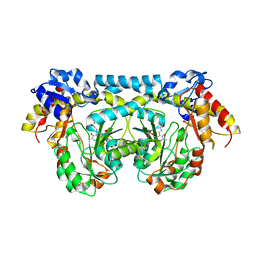 | |
4L9R
 
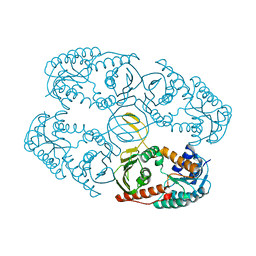 | |
4L8V
 
 | | Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP | | Descriptor: | 1,2-ETHANEDIOL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Converting NAD-Specific Inositol Dehydrogenase to an Efficient NADP-Selective Catalyst, with a Surprising Twist.
Biochemistry, 52, 2013
|
|
4K2I
 
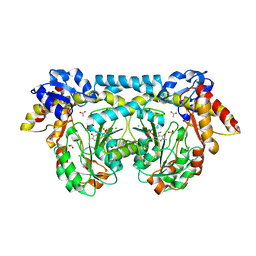 | | Crystal structure of ntda from bacillus subtilis with bound cofactor pmp | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4LIS
 
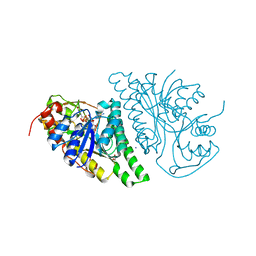 | | Crystal Structure of UDP-galactose-4-epimerase from Aspergillus nidulans | | Descriptor: | GLYCEROL, IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Ko, J, Sheoran, I, Kaminskyj, S.G.W, Sanders, D.A.R. | | Deposit date: | 2013-07-03 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidation of Substrate Specificity in Aspergillus nidulans UDP-Galactose-4-Epimerase.
Plos One, 8, 2013
|
|
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
6TZU
 
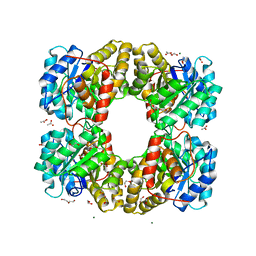 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
1AIU
 
 | | HUMAN THIOREDOXIN (D60N MUTANT, REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Andersen, J.F, Gasdaska, J.R, Sanders, D.A.R, Weichsel, A, Powis, G, Montfort, W.R. | | Deposit date: | 1997-04-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 --> asparagine mutant.
Biochemistry, 36, 1997
|
|
4MO2
 
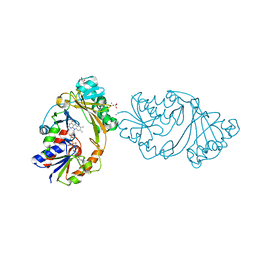 | | Crystal Structure of UDP-N-acetylgalactopyranose mutase from Campylobacter jejuni | | Descriptor: | AMMONIUM ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Protsko, C, Poulin, M.B, Lowary, T.L, Sanders, D.A.R. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of a UDP-GalNAc Pyranose-Furanose Mutase: A Potential Therapeutic Target for Campylobacter jejuni Infections.
Chembiochem, 15, 2014
|
|
1AUC
 
 | | HUMAN THIOREDOXIN (OXIDIZED WITH DIAMIDE) | | Descriptor: | THIOREDOXIN | | Authors: | Anderson, J.F, Sanders, D.A.R, Gasdaska, J, Weichsel, A, Powis, G, Montfort, W.R. | | Deposit date: | 1997-08-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 --> asparagine mutant.
Biochemistry, 36, 1997
|
|
4MIO
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD(H) and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD(H) and myo-inositol
To be Published
|
|
4MIE
 
 | |
4MKZ
 
 | |
