1QGI
 
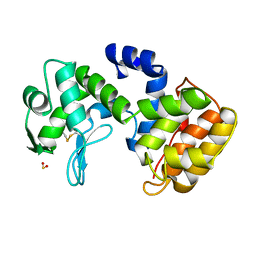 | | CHITOSANASE FROM BACILLUS CIRCULANS | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | Authors: | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
3WXC
 
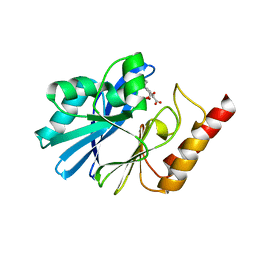 | | Crystal Structure of IMP-1 metallo-beta-lactamase complexed with a 3-aminophtalic acid inhibitor | | Descriptor: | 3-(4-hydroxypiperidin-1-yl)benzene-1,2-dicarboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Saito, J, Watanabe, T, Yamada, M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic analysis of IMP-1 metallo-beta-lactamase complexed with a 3-aminophthalic acid derivative, structure-based drug design, and synthesis of 3,6-disubstituted phthalic acid derivative inhibitors
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2Z6J
 
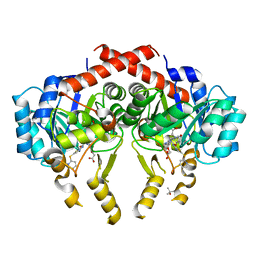 | | Crystal Structure of S. pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabK) in Complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-(2-((3-(5-(PYRIDIN-2-YLTHIO)THIAZOL-2-YL)UREIDO)METHYL)-1H-IMIDAZOL-4-YL)PHENOXY)ACETIC ACID, CALCIUM ION, ... | | Authors: | Saito, J, Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-08-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-acyl carrier protein reductase (FabK) from Streptococcus pneumoniae reveals the binding mode of an inhibitor.
Protein Sci., 17, 2008
|
|
2Z6I
 
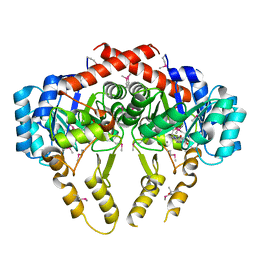 | | Crystal Structure of S. pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabK) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Saito, J, Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-08-01 | | Release date: | 2008-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of enoyl-acyl carrier protein reductase (FabK) from Streptococcus pneumoniae reveals the binding mode of an inhibitor.
Protein Sci., 17, 2008
|
|
5HWA
 
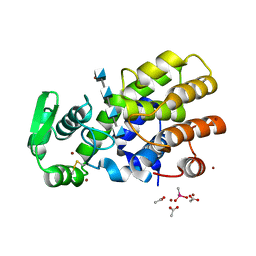 | | Crystal Structure of MH-K1 chitosanase in substrate-bound form | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Suzuki, M, Saito, A, Ando, A, Miki, K, Saito, J. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Biomed.Biochim.Acta, 1868, 2024
|
|
8GNE
 
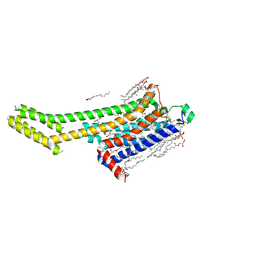 | | Crystal structure of human adenosine A2A receptor in complex with an insurmountable inverse agonist, KW-6356. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
8GNG
 
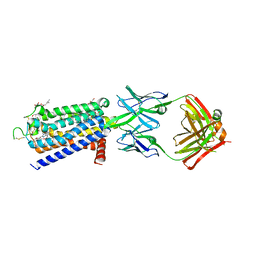 | | Crystal structure of human adenosine A2A receptor in complex with istradefylline. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 8-[(~{E})-2-(3,4-dimethoxyphenyl)ethenyl]-1,3-diethyl-7-methyl-purine-2,6-dione, Adenosine receptor A2a, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
5ETY
 
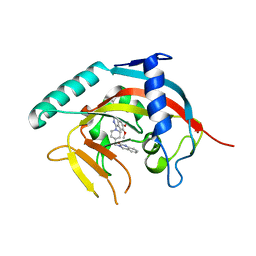 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1J
 
 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
2VEE
 
 | | Structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2VEB
 
 | | High resolution structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | GLYCEROL, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
7XRZ
 
 | | Crystal structure of BRIL and SRP2070_Fab complex | | Descriptor: | IGG HEAVY CHAIN, IGG LIGHT CHAIN, Soluble cytochrome b562 | | Authors: | Suzuki, M, Miyagi, H, Yasunaga, M, Asada, H, Iwata, S, Saito, J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into an anti-BRIL Fab as a G-protein-coupled receptor crystallization chaperone.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7C6B
 
 | | Crystal structure of Ago2 MID domain in complex with 6-(3-(2-carboxyethyl)phenyl)purine riboside monophosphate | | Descriptor: | 3-[3-[9-[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]purin-6-yl]phenyl]propanoic acid, PHOSPHATE ION, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
6K0T
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-17 | | Descriptor: | 3-[(1~{E})-1-[8-[(8-chloranyl-2-cyclopropyl-imidazo[1,2-a]pyridin-3-yl)methyl]-3-fluoranyl-6~{H}-benzo[c][1]benzoxepin-11-ylidene]ethyl]-4~{H}-1,2,4-oxadiazol-5-one, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Suzuki, M, Yamamoto, K, Takahashi, Y, Saito, J. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of a novel class of peroxisome proliferator-activated receptor (PPAR) gamma ligands as an anticancer agent with a unique binding mode based on a non-thiazolidinedione scaffold.
Bioorg.Med.Chem., 27, 2019
|
|
7D7U
 
 | | Crystal structure of Ago2 MID domain in complex with 8-Br-adenosin-5'-monophosphate | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-10-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
7CE4
 
 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
7CMM
 
 | | Crystal structure of TEAD1-YBD in complex with K-975 | | Descriptor: | N-[3-(4-chloranylphenoxy)-4-methyl-phenyl]propanamide, Transcriptional enhancer factor TEF-1 | | Authors: | Tsuji, Y, Suzuki, M, Yasunaga, M, Hamguchi, K, Saito, J. | | Deposit date: | 2020-07-28 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The novel potent TEAD inhibitor, K-975, inhibits YAP1/TAZ-TEAD protein-protein interactions and exerts an anti-tumor effect on malignant pleural mesothelioma.
Am J Cancer Res, 10, 2020
|
|
7C6A
 
 | | Crystal structure of AT2R-BRIL and SRP2070_Fab complex | | Descriptor: | IgG Light Chain, IgG heavy chain, SAR1, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
7C61
 
 | | Crystal structure of 5-HT1B-BRIL and SRP2070_Fab complex | | Descriptor: | 5-hydroxytryptamine receptor 1B,Soluble cytochrome b562,5-hydroxytryptamine receptor 1B, Ergotamine, IGG HEAVY CHAIN, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
4E2O
 
 | | Crystal structure of alpha-amylase from Geobacillus thermoleovorans, GTA, complexed with acarbose | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Mok, S.C, Teh, A.H, Saito, J.A, Najimudin, N, Alam, M. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans.
Enzyme.Microb.Technol., 53, 2013
|
|
3VOT
 
 | | Crystal structure of L-amino acid ligase from Bacillus licheniformis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, M, Takahashi, Y, Noguchi, A, Arai, T, Yagasaki, M, Kino, K, Saito, J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of L-amino-acid ligase from Bacillus licheniformis
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3AVE
 
 | | Crystal Structure of the Fucosylated Fc Fragment from Human Immunoglobulin G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, ZINC ION | | Authors: | Matsumiya, S, Yamaguchi, Y, Saito, J, Nagano, M, Sasakawa, H, Otaki, S, Satoh, M, Shitara, K, Kato, K. | | Deposit date: | 2011-03-04 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Corrigendum to "Structural Comparison of Fucosylated and Nonfucosylated Fc Fragments of Human Immunoglobulin G1" [J. Mol. Biol. 386/3 (2007) 767-779]
J.Mol.Biol., 408, 2011
|
|
2D05
 
 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
