4DCA
 
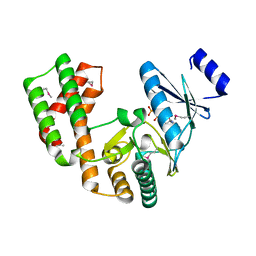 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound
TO BE PUBLISHED
|
|
7MWA
 
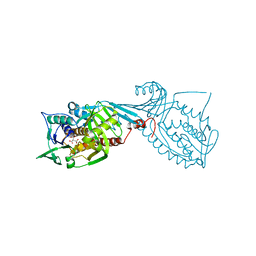 | | Crystal structure of 2-octaprenyl-6-methoxyphenol hydroxylase UbiH from Acinetobacter baumannii, apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-polyprenyl-6-methoxyphenol 4-hydroxylase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UbiH from Acinetobacter baumannii
To Be Published
|
|
3BRQ
 
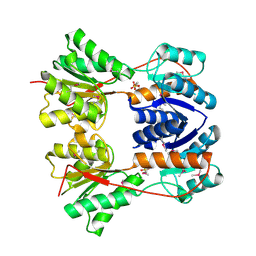 | | Crystal structure of the Escherichia coli transcriptional repressor ascG | | Descriptor: | HTH-type transcriptional regulator ascG, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the E. coli transcriptional repressor ascG.
To be Published
|
|
5VYV
 
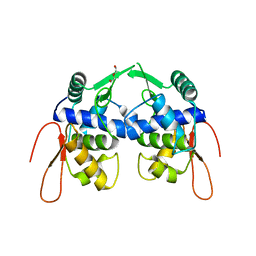 | | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli | | Descriptor: | DI(HYDROXYETHYL)ETHER, UPF0502 protein YceH | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
4DGT
 
 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 crystallized with magnesium formate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
7JM0
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme | | Descriptor: | Aminocyclitol acetyltransferase ApmA, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme
To Be Published
|
|
3C0U
 
 | | Crystal structure of E.coli yaeQ protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein yaeQ | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli yaeQ protein.
To be Published
|
|
5KOL
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECK1530/EC0983 from Escherichia coli | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-30 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | To be published
To Be Published
|
|
5VKW
 
 | | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans | | Descriptor: | Adenylosuccinate lyase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans
To Be Published
|
|
2LOJ
 
 | | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273 | | Descriptor: | Putative cytoplasmic protein | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273
To be Published
|
|
3C9H
 
 | | Crystal structure of the substrate binding protein of the ABC transporter from Agrobacterium tumefaciens | | Descriptor: | ABC transporter, substrate binding protein, CITRIC ACID, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the substrate binding protein of the ABC transporter from Agrobacterium tumefaciens.
To be Published
|
|
6E0S
 
 | | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library | | Descriptor: | GLYCEROL, MEM-A1, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Lau, C, Topp, E, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library
To Be Published
|
|
4EXL
 
 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphate-binding protein pstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A
To be Published
|
|
3CTV
 
 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
3TYS
 
 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3UDU
 
 | | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, CHLORIDE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Grimshaw, S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni
To be Published
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
2M46
 
 | | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841 | | Descriptor: | Arsenate reductase, putative | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841
To be Published
|
|
4EJ7
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside 3'-phosphotransferase AphA1-IAB, CALCIUM ION, ... | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Evdokimova, E, Egorova, O, Di Leo, R, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4ECL
 
 | | Crystal structure of the cytoplasmic domain of vancomycin resistance serine racemase VanTg | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine racemase | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Cosme, J, Di Leo, R, Krishnamoorthy, M, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structural and Functional Adaptation of Vancomycin Resistance VanT Serine Racemases.
MBio, 6, 2015
|
|
4DE4
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-Id/APH(2")-IVa in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APH(2")-Id | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
3BIG
 
 | | Crystal structure of the fructose-1,6-bisphosphatase GlpX from E.coli in complex with inorganic phosphate | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3BQY
 
 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
4EDP
 
 | | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124 | | Descriptor: | ABC transporter, substrate-binding protein, ACETATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124
To be Published
|
|
