3I9S
 
 | | Structure from the mobile metagenome of V.cholerae. Integron cassette protein VCH_CASS6 | | Descriptor: | CHLORIDE ION, Integron cassette protein, SULFATE ION | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-13 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure from the mobile metagenome of V.cholerae. Integron cassette protein VCH_CASS6
To be Published
|
|
3O12
 
 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
3IAH
 
 | | Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate. | | Descriptor: | ACETATE ION, CHLORIDE ION, ETHANOL, ... | | Authors: | Minasov, G, Halavaty, A, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate.
TO BE PUBLISHED
|
|
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
3KBO
 
 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3OBB
 
 | | Crystal structure of a possible 3-hydroxyisobutyrate Dehydrogenase from pseudomonas aeruginosa pao1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Yakunin, A.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3K6R
 
 | | CRYSTAL STRUCTURE OF putative transferase PH0793 FROM PYROCOCCUS HORIKOSHII | | Descriptor: | putative transferase PH0793 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-09 | | Release date: | 2009-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Protein Ph0793 from Pyrococcus Horikoshii
To be Published
|
|
3OI7
 
 | | Structure of the structure of the H13A mutant of Ykr043C in complex with sedoheptulose-1,7-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,7-di-O-phosphono-beta-D-altro-hept-2-ulofuranose, GLYCEROL, ... | | Authors: | Singer, A.U, Xu, X, Dong, A, Cui, H, Clasquin, M.F, Caudy, A.A, Edwards, A.M, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Riboneogenesis in yeast.
Cell(Cambridge,Mass.), 145, 2011
|
|
3OSU
 
 | | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Edwards, A, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-09 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus
To be Published
|
|
3KWP
 
 | | Crystal structure of putative methyltransferase from Lactobacillus brevis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Predicted methyltransferase | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-01 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of putative methyltransferase from Lactobacillus brevis
To be Published
|
|
3P48
 
 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3K28
 
 | | Crystal Structure of a glutamate-1-semialdehyde aminotransferase from Bacillus anthracis with bound Pyridoxal 5'Phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glutamate-1-semialdehyde 2,1-aminomutase 2, ... | | Authors: | Sharma, S.S, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2010-01-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a glutamate-1-semialdehyde aminotransferase from Bacillus anthracis with bound Pyridoxal 5'Phosphate
To be Published
|
|
3K6C
 
 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
3KYE
 
 | | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis | | Descriptor: | Roadblock/LC7 domain, Robl_LC7 | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-05 | | Release date: | 2009-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis
To be Published
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3K67
 
 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin
To Be Published
|
|
3K3S
 
 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
3H0P
 
 | | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium. | | Descriptor: | S-malonyltransferase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3PFC
 
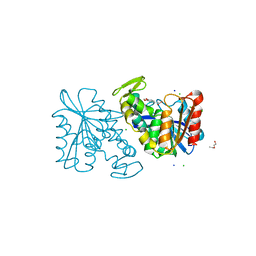 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 S106A mutant in complex with ferulic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2010-10-28 | | Release date: | 2011-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
3NKD
 
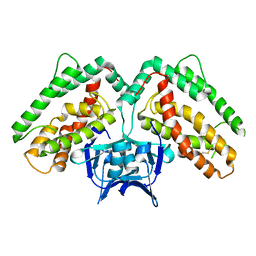 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
3LED
 
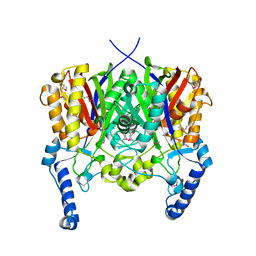 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009 | | Descriptor: | 3-oxoacyl-acyl carrier protein synthase III, FORMIC ACID | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-14 | | Release date: | 2010-01-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009
To be Published
|
|
3N08
 
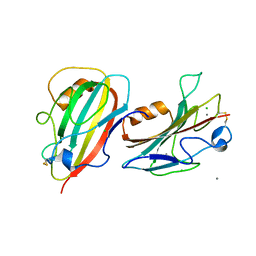 | | Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative PhosphatidylEthanolamine-Binding Protein (PEBP) | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 Angstrom Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX
To be Published
|
|
3N3T
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
