6PY8
 
 | | Crystal structure of the RBPJ-NOTCH1-NRARP ternary complex bound to DNA | | Descriptor: | DNA, Neurogenic locus notch homolog protein 1, Notch-regulated ankyrin repeat-containing protein, ... | | Authors: | Jarrett, S.M, Seegar, T.C.M, Blacklow, S.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Sci.Signal., 12, 2019
|
|
6PVU
 
 | | RNase A in complex with hexametaphosphate | | Descriptor: | 2,4,6,8,10,12-hexahydroxy-2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~,10lambda~5~,12lambda~5~-cyclohexaphosphoxane-2,4,6,8,10,12-hexone, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PVW
 
 | | RNase A in complex with cp4pA | | Descriptor: | 2,4,6,8-tetrahydroxy-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocane-2,4,6,8-tetrone, 5'-O-[(R)-hydroxy{[(4R,8S)-4,6,8-trihydroxy-2,4,6,8-tetraoxo-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocan-2-yl]oxy}phosphoryl]adenosine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
8A40
 
 | | Structure of mammalian Pol II-TFIIS elongation complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Farnung, L, Ochmann, M, Garg, G, Vos, S.M, Cramer, P. | | Deposit date: | 2022-06-09 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a backtracked hexasomal intermediate of nucleosome transcription.
Mol.Cell, 82, 2022
|
|
8SLO
 
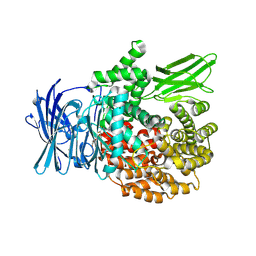 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
2FTB
 
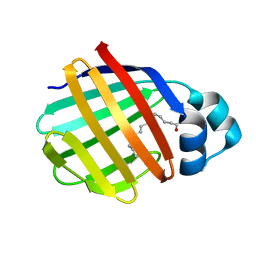 | | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to oleic acid | | Descriptor: | Fatty acid-binding protein 2, liver, OLEIC ACID | | Authors: | Capaldi, S, Guariento, M, Perduca, M, Di Pietro, S.M, Santome, J.A, Monaco, H.L. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to cholic and oleic acid
Proteins, 64, 2006
|
|
7RAR
 
 | |
7RJ2
 
 | |
7RJ4
 
 | |
7RHN
 
 | |
7RHM
 
 | |
7RMJ
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor (S)-N-(1-(3-(4-chloro-3-(methylsulfonamido)-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl)-6-(3-methyl-3-(methylsulfonyl)but-1-yn-1-yl)pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CAPSID PROTEIN P24, CHLORIDE ION, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and Mechanistic Bases of Viral Resistance to HIV-1 Capsid Inhibitor Lenacapavir.
Mbio, 13, 2022
|
|
7RMM
 
 | |
7RAO
 
 | |
7S2N
 
 | |
7S2H
 
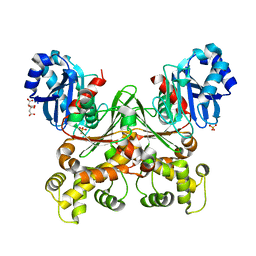 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
7S2P
 
 | | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, SULFATE ION | | Authors: | Carey, S.M, Nettles, R.B, Daneshian, L, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN
To Be Published
|
|
7S04
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (IspC) from Acinetobacter baumannii in complex with FR900098, NADPH, and a magnesium ion | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, MAGNESIUM ION, ... | | Authors: | Noble, S.M, Ball, H.S, Couch, R.D. | | Deposit date: | 2021-08-28 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Characterization and Inhibition of 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase: A Promising Drug Target in Acinetobacter baumannii and Klebsiella pneumoniae .
Acs Infect Dis., 7, 2021
|
|
7QPU
 
 | | Botulinum neurotoxin A5 cell binding domain in complex with GM1b oligosaccharide | | Descriptor: | Botulinum neurotoxin sub-type A5, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
7QPT
 
 | | Botulinum neurotoxin A4 cell binding domain in complex with GD1a oligosaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type A | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mojanaga, O.O. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
2FT9
 
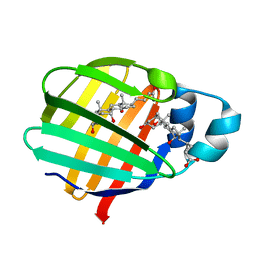 | | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to cholic acid | | Descriptor: | CHOLIC ACID, Fatty acid-binding protein 2, liver | | Authors: | Capaldi, S, Guariento, M, Perduca, M, Di Pietro, S.M, Santome, J.A, Monaco, H.L. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to cholic and oleic acid
Proteins, 64, 2006
|
|
7SB2
 
 | |
7SAT
 
 | |
7SAZ
 
 | |
7SAU
 
 | |
