4ASG
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, NICKEL (II) ION, [(3R,5S,6R,7R,11S,12Z,14E)-5,11,21-trimethoxy-3,7,9,15-tetramethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-19-phenyl-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
3O3H
 
 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and manganese ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
4AS9
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, [(3R,5S,6R,7R,10R,11S,12E)-5,11,21-trimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4ASF
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | (8S,9R,13R,14S,16R)-21-(furan-2-yl)-13-hydroxy-8,14,19-trimethoxy-16-methyl-4,10,12-trimethylidene-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),18-dien-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, NICKEL (II) ION | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4AVA
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4ALF
 
 | | Pseudomonas fluorescens PhoX in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALKALINE PHOSPHATASE PHOX, CALCIUM ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
8GKC
 
 | |
4A38
 
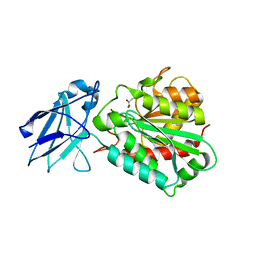 | | METALLO-CARBOXYPEPTIDASE FROM PSEUDOMONAS AUREGINOSA IN COMPLEX WITH L-BENZYLSUCCINIC ACID | | Descriptor: | L-BENZYLSUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4AE4
 
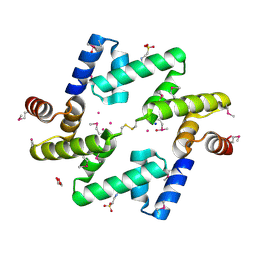 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
4A9X
 
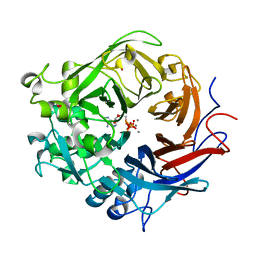 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MU-OXO-DIIRON, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
4C7I
 
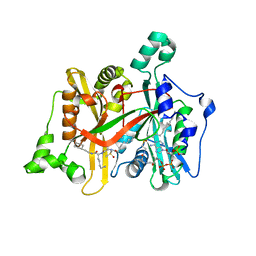 | | Leismania major N-myristoyltransferase in complex with a peptidomimetic (-OH) molecule | | Descriptor: | DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Olaleye, T.O, Brannigan, J.A, Goncalves, V, Roberts, S.M, Leatherbarrow, R.J, Wilkinson, A.J, Tate, E.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptidomimetic Inhibitors of N-Myristoyltransferase from Human Malaria and Leishmaniasis Parasites.
Org.Biomol.Chem., 12, 2014
|
|
4CBZ
 
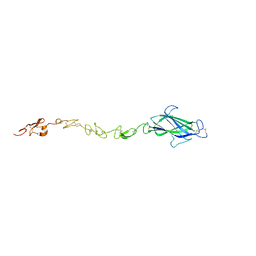 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN JAGGED-1, alpha-L-fucopyranose | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4CC1
 
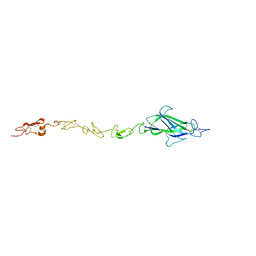 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4BFE
 
 | | Structure of the extracellular portion of mouse CD200RLa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELL SURFACE GLYCOPROTEIN CD200 RECEPTOR 4, CYSTEINE, ... | | Authors: | Hatherley, D, Lea, S.M, Johnson, S, Barclay, A.N. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Cd200/Cd200 Receptor Family and Implications for Topology, Regulation, and Evolution
Structure, 21, 2013
|
|
4B75
 
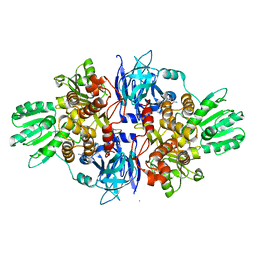 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4ASA
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CHLORIDE ION, [(3R,5S,6R,7R,12E)-5,11-dimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-21-(prop-2-enylamino)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
3OHW
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 721-860) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209E | | Descriptor: | Phycobilisome LCM core-membrane linker polypeptide | | Authors: | Kuzin, A, Su, M, Lew, S, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-09-01 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209E
To be Published
|
|
4BF3
 
 | | ErpC, a member of the complement regulator acquiring family of surface proteins from Borrelia burgdorfei, possesses an architecture previously unseen in this protein family. | | Descriptor: | 1,2-ETHANEDIOL, ERPC | | Authors: | Caesar, J.J.E, Johnson, S, Kraiczy, P, Lea, S.M. | | Deposit date: | 2013-03-14 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Erpc, a Member of the Complement Regulator-Acquiring Family of Surface Proteins from Borrelia Burgdorferi, Possesses an Architecture Previously Unseen in This Protein Family.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AMF
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
3P2S
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in an open conformation | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P3Q
 
 | | Crystal Structure of MmoQ Response regulator from Methylococcus capsulatus str. Bath at the resolution 2.4A, Northeast Structural Genomics Consortium Target McR175M | | Descriptor: | MmoQ, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Abashidze, M, Seetharaman, J, Janjua, J, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-10-05 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR175M
To be Published
|
|
4BUP
 
 | | A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases | | Descriptor: | GLYCEROL, HISTONE-LYSINE N-METHYLTRANSFERASE SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Southall, S.M, Cronin, N.B, Wilson, J.R. | | Deposit date: | 2013-06-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.166 Å) | | Cite: | A Novel Route to Product Specificity in the Suv4-20 Family of Histone H4K20 Methyltransferases.
Nucleic Acids Res., 42, 2014
|
|
4B74
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-[(2-ammonioethyl)amino]-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function
Nat.Chem.Biol., 8, 2012
|
|
4CE3
 
 | | Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. | | Descriptor: | 13-Chloro-14,16-dihydroxy-2-methyl-2,3,4,5,9,10-hexahydrobenz[c][1]azacyclotetradecine-1,11(8H,12H)-dione, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Parry-Morris, S, Prodromou, C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Synthesis of Macrolactam Analogues of Radicicol and Their Binding to Heat Shock Protein Hsp90.
Org.Biomol.Chem., 12, 2014
|
|
1ZTR
 
 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
