4IL3
 
 | | Crystal Structure of S. mikatae Ste24p | | Descriptor: | Ste24p, ZINC ION | | Authors: | Pryor Jr, E.E, Horanyi, P.S, Clark, K, Fedoriw, N, Connelly, S.M, Koszelak-Rosenblum, M, Zhu, G, Malkowski, M.G, Dumont, M.E, Wiener, M.C, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structure of the integral membrane protein CAAX protease Ste24p.
Science, 339, 2013
|
|
4IMN
 
 | | Crystal structure of wild type human Lipocalin PGDS bound with PEG MME 2000 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lipocalin-type prostaglandin-D synthase | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4IPM
 
 | | Crystal structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with thiocellobiose | | Descriptor: | ACETATE ION, CALCIUM ION, GH7 family protein, ... | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1HFO
 
 | |
4IPX
 
 | | Analyzing the visible conformational substates of the FK506 binding protein FKBP12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysing the visible conformational substates of the FK506-binding protein FKBP12.
Biochem.J., 453, 2013
|
|
4IQ2
 
 | | P21 crystal form of FKBP12.6 | | Descriptor: | MALONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IQC
 
 | | P3121 crystal form of FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2AAC
 
 | |
6X4H
 
 | | Sortilin-Progranulin Interaction With Compound 24 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-N-(6-phenoxypyridine-3-carbonyl)-L-leucine, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M, Klein, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WVQ
 
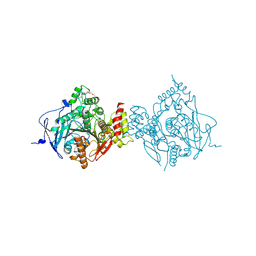 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GP | | Descriptor: | (1S)-2,2-dimethylcyclopentyl (R)-methylphosphinate, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WVC
 
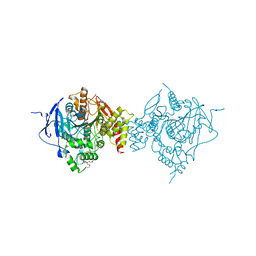 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GD | | Descriptor: | (1S)-2,2-dimethylcyclopentyl (R)-methylphosphinate, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WUV
 
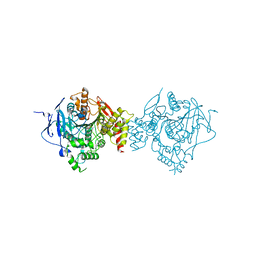 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WVP
 
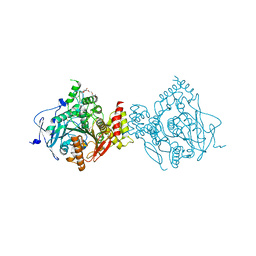 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GF | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WV1
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GB and HI-6 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WUY
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GA and HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WUZ
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by GB | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6WVO
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GD and HI-6 | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
6KL6
 
 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
8CUR
 
 | | Crystal structure of Cdk2 in complex with Cyclin A inhibitor 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid | | Descriptor: | 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid, Cyclin-dependent kinase 2 | | Authors: | Tripathi, S.M, Tambo, C.S, Kiss, G, Rubin, S.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biolayer Interferometry Assay for Cyclin-Dependent Kinase-Cyclin Association Reveals Diverse Effects of Cdk2 Inhibitors on Cyclin Binding Kinetics.
Acs Chem.Biol., 18, 2023
|
|
6KL2
 
 | |
6XKR
 
 | | Structure of Sasanlimab Fab in complex with PD-1 | | Descriptor: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | Authors: | Kimberlin, C.R, Chin, S.M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
2X1K
 
 | |
6XF1
 
 | |
1GNP
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID 3'-O-(N-METHYLANTHRANILOYL-2'-DEOXYGUANYLATE ESTER | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
6XR8
 
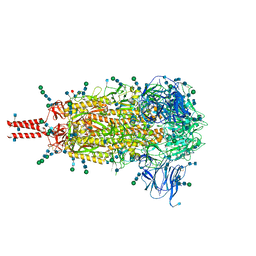 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
