2WW4
 
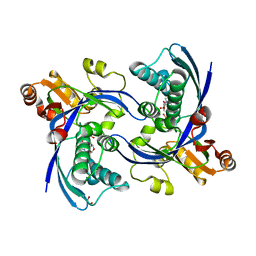 | | a triclinic crystal form of E. coli 4-diphosphocytidyl-2C-methyl-D- erythritol kinase | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL | | Authors: | Kalinowska-Tluscik, J, Miallau, L, Gabrielsen, M, Leonard, G.A, McSweeney, S.M, Hunter, W.N. | | Deposit date: | 2009-10-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triclinic Crystal Form of Escherichia Coli 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase and Reassessment of the Quaternary Structure.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1HEJ
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
3T60
 
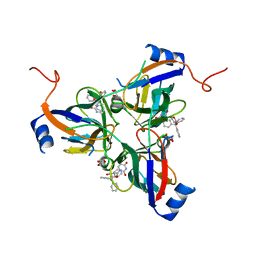 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-(tritylamino)uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
1HEH
 
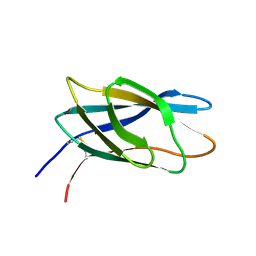 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1H8T
 
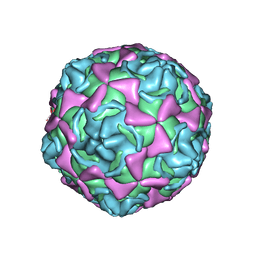 | | Echovirus 11 | | Descriptor: | 12-AMINO-DODECANOIC ACID, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Stuart, A, McKee, T, Williams, P.A, Harley, C, Stuart, D.I, Brown, T.D.K, Lea, S.M. | | Deposit date: | 2001-02-15 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Determination of the Structure of a Decay Accelerating Factor-Binding Clinical Isolate of Echovirus 11 Allows Mapping of Mutants with Altered Receptor Requirements for Infection
J.Virol., 76, 2002
|
|
1H8F
 
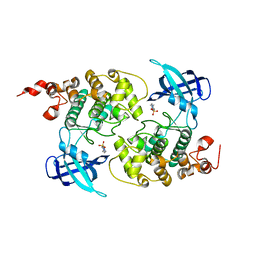 | | Glycogen Synthase Kinase 3 beta. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Dajani, R, Pearl, L.H, Roe, S.M. | | Deposit date: | 2001-02-05 | | Release date: | 2002-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glycogen Synthase Kinase 3Beta . Structural Basis for Phosphate-Primed Substrate Specificity and Autoinhibition
Cell(Cambridge,Mass.), 105, 2001
|
|
1GS0
 
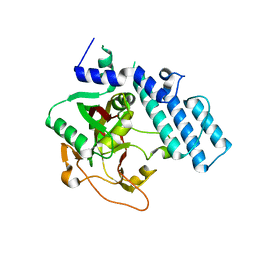 | |
3T64
 
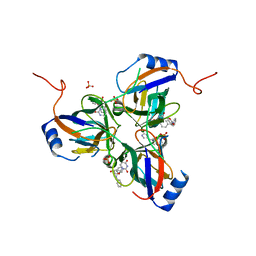 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-[(diphenylmethyl)amino]uridine, 5'-(BENZHYDRYLAMINO)-2',5'-DIDEOXYURIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
6UZT
 
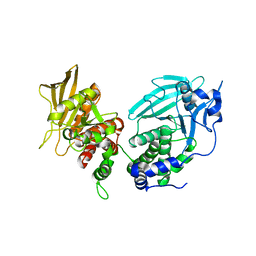 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
5FPX
 
 | | The structure of KdgF from Yersinia enterocolitica. | | Descriptor: | NICKEL (II) ION, PECTIN DEGRADATION PROTEIN, PEPTIDE | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3C37
 
 | | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A. Northeast Structural Genomics Consortium target GsR143A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidase, M48 family, ... | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Vorobiev, S.M, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
To be Published
|
|
2VVK
 
 | | Grb2 SH3C (1) | | Descriptor: | GLYCEROL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
5FQ0
 
 | | The structure of KdgF from Halomonas sp. | | Descriptor: | CITRATE ANION, KDGF, NICKEL (II) ION, ... | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1FST
 
 | | CRYSTAL STRUCTURE OF TRUNCATED HUMAN RHOGDI TRIPLE MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6SLH
 
 | | Conformational flexibility within the small domain of human serine racemase. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Serine racemase, ... | | Authors: | Koulouris, C.R, Bax, B, Atack, J, Roe, S.M. | | Deposit date: | 2019-08-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Conformational flexibility within the small domain of human serine racemase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2X04
 
 | | Crystal structure of the PABC-TNRC6C complex | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1, SULFATE ION, TRINUCLEOTIDE REPEAT-CONTAINING GENE 6C PROTEIN | | Authors: | Jinek, M, Fabian, M.R, Coyle, S.M, Sonenberg, N, Doudna, J.A. | | Deposit date: | 2009-12-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Insights Into the Human Gw182-Pabc Interaction in Microrna-Mediated Deadenylation
Nat.Struct.Mol.Biol., 17, 2010
|
|
5FDB
 
 | |
2WYN
 
 | | Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue | | Descriptor: | (1R,2R,3R,6R,7R,7AR)-3,7-BIS(HYDROXYMETHYL)HEXAHYDRO-1H-PYRROLIZINE-1,2,6-TRIOL, CALCIUM ION, PERIPLASMIC TREHALASE, ... | | Authors: | Gloster, T.M, Roberts, S.M, Davies, G.J, Cardona, F, Goti, A, Parmeggiani, C, Parenti, P, Fusi, P, Forcella, M, Cipolla, L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Casuarine-6-O-alpha-D-glucoside and its analogues are tight binding inhibitors of insect and bacterial trehalases.
Chem.Commun.(Camb.), 46, 2010
|
|
3UT0
 
 | | Crystal structure of exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-11-24 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 2011
|
|
5FYG
 
 | | Structure of CYP153A from Marinobacter aquaeolei in complex with hydroxydodecanoic acid | | Descriptor: | 12-HYDROXYDODECANOIC ACID, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh-Azari, H.-R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
3D5N
 
 | | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125. | | Descriptor: | Q97W15_SULSO | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Lee, D, Foote, R.E, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-16 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125.
To be Published
|
|
6UST
 
 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
7NPI
 
 | | Crystal structure of Mindy2 (C266A) in complex with Lys48-linked penta-ubiquitin (K48-Ub5) | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, SODIUM ION, ... | | Authors: | Lange, S.M, Armstrong, L.A, Kulathu, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
2XRC
 
 | | Human complement factor I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HUMAN COMPLEMENT FACTOR I | | Authors: | Roversi, P, Johnson, S, Lea, S.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Basis for Complement Factor I Control and its Disease-Associated Sequence Polymorphisms.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
