2XX4
 
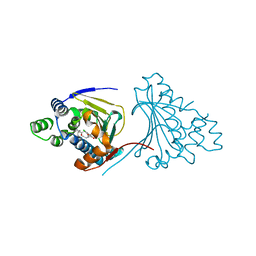 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (E)-ETHYL 13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDROBENZO[C][1]AZACYCLOTETRADECINE-10-CARBOXYLATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
3USZ
 
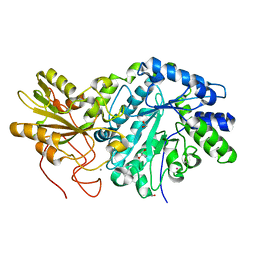 | | Crystal structure of truncated exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-11-24 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 2011
|
|
5FYF
 
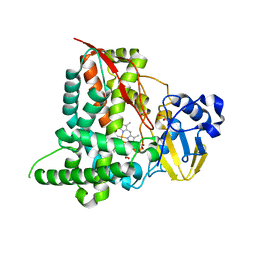 | | Structure of CYP153A from Marinobacter aquaeolei | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh Azari, H.R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
5FMW
 
 | | The poly-C9 component of the Complement Membrane Attack Complex | | Descriptor: | POLYC9 | | Authors: | Dudkina, N.V, Spicer, B.A, Reboul, C.F, Conroy, P.J, Lukoyanova, N, Elmlund, H, Law, R.H.P, Ekkel, S.M, Kondos, S.C, Goode, R.J.A, Ramm, G, Whisstock, J.C, Saibil, H.R, Dunstone, M.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the Poly-C9 Component of the Complement Membrane Attack Complex
Nat.Commun., 7, 2016
|
|
3V2Y
 
 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 2.80A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera (E.C.3.2.1.17), ... | | Authors: | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
6TC5
 
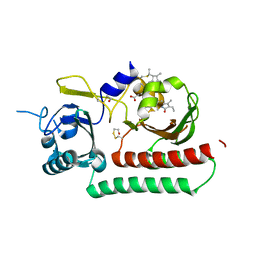 | | Phytochromobilin-adducted PAS-GAF bidomain of Sorghum bicolor phyB | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, BETA-MERCAPTOETHANOL, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
1GML
 
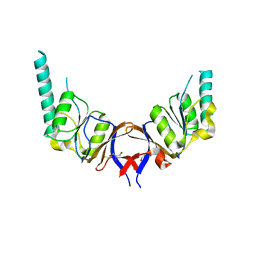 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GN1
 
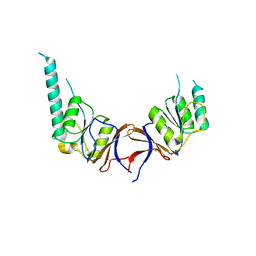 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
2YI6
 
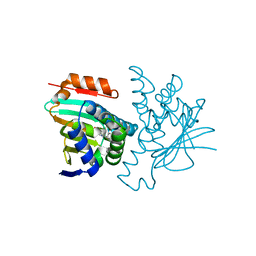 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | Descriptor: | 4-[5-(4-ETHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL]-6-ETHYLBENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-05-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
2YI7
 
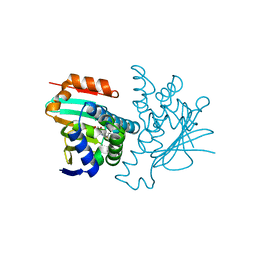 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | Descriptor: | 4-CHLORO-6-[5-(4-ETHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-05-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
6TNZ
 
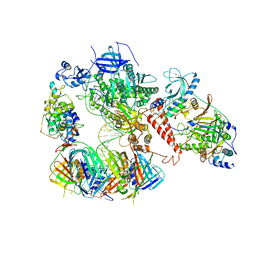 | | Human polymerase delta-FEN1-PCNA toolbelt | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6TRE
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 32-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the bacterial flagellar rotor MS-ring: a minimum inventory/maximum diversity system.
To Be Published
|
|
1GYE
 
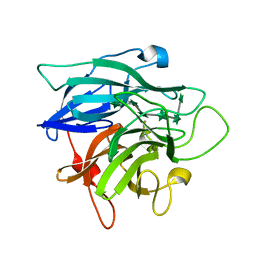 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase complexed with Arabinohexaose | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1GNQ
 
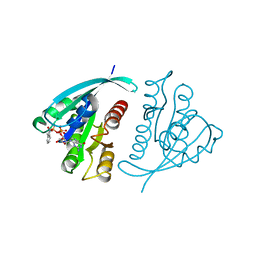 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1HQ5
 
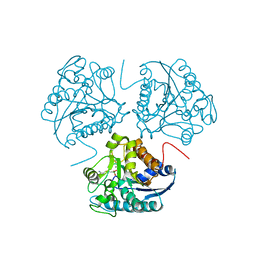 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
2XZ9
 
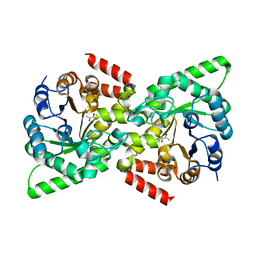 | | CRYSTAL STRUCTURE FROM THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA), PYRUVIC ACID | | Authors: | Oberholzer, A.E, Waltersperger, S.M, Schneider, P, Baumann, U, Erni, B. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
6TED
 
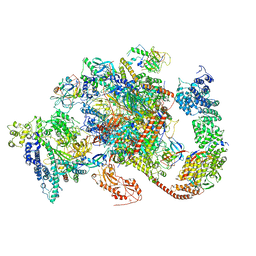 | | Structure of complete, activated transcription complex Pol II-DSIF-PAF-SPT6 uncovers allosteric elongation activation by RTF1 | | Descriptor: | DNA (37-MER), DNA-directed RNA polymerase II subunit RPB9, DNA-directed RNA polymerase subunit, ... | | Authors: | Vos, S.M, Farnung, L, Cramer, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2XGA
 
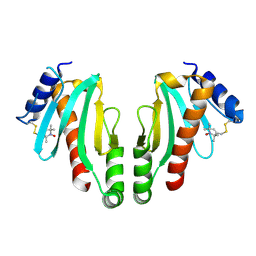 | | MTSL spin-labelled Shigella Flexneri Spa15 | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, SURFACE PRESENTATION OF ANTIGENS PROTEIN SPAK | | Authors: | Lillington, J.E.D, Johnson, S, Lea, S.M. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shigella Flexneri Spa15 Crystal Structure Verified in Solution by Double Electron Electron Resonance.
J.Mol.Biol., 405, 2011
|
|
7JJK
 
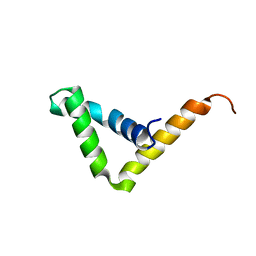 | | Crystal structure of SOX30 | | Descriptor: | Transcription factor SOX-30 | | Authors: | Ghafoori, S.M, Forwood, J.K. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of SOX30 at 1.40 Angstroms resolution
To Be Published
|
|
5EBE
 
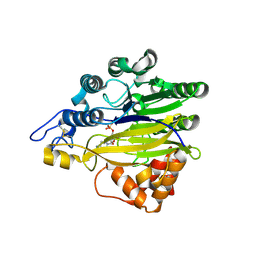 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with 5' CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-O-phosphono-beta-D-ribofuranose, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
3B57
 
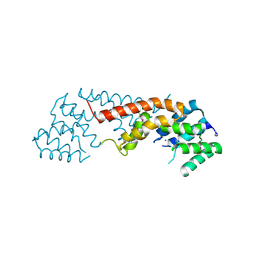 | | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua. Northeast Structural Consortium target LkR65 | | Descriptor: | Lin1889 protein, MAGNESIUM ION | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Benach, J, Forouhar, F, Vorobiev, S.M, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua.
To be Published
|
|
2XL2
 
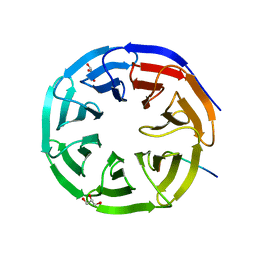 | |
2XD6
 
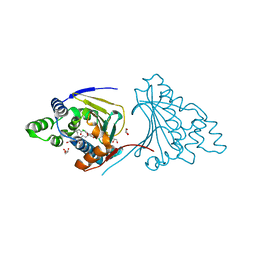 | | Hsp90 complexed with a resorcylic acid macrolactone. | | Descriptor: | (5Z)-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-3,4,7,8,9,10,11,12-OCTAHYDRO-1H-2-BENZOXACYCLOTETRADECINE-6-CARBALDEHYDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H, Moody, C.J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of Hsp90 with Resorcylic Acid Macrolactones. Synthesis and Binding Studies.
Chemistry, 16, 2010
|
|
1HJ9
 
 | | Atomic resolution structures of trypsin provide insight into structural radiation damage | | Descriptor: | ANILINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, McSweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-10 | | Release date: | 2002-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
