4LOA
 
 | | X-ray structure of the de-novo design amidase at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target OR398 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, De-novo design amidase | | Authors: | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Khersonsky, O, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR398
To be Published
|
|
3EQE
 
 | | Crystal structure of the YubC protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR112. | | Descriptor: | FE (III) ION, Putative cystein dioxygenase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Benach, J, Forouhar, F, Clayton, G, Cooper, B, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the YubC protein from Bacillus subtilis.
To be Published
|
|
3QSZ
 
 | | Crystal Structure of the STAR-related lipid transfer protein (fragment 25-204) from Xanthomonas axonopodis at the resolution 2.4A, Northeast Structural Genomics Consortium Target XaR342 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, STAR-related lipid transfer protein, ... | | Authors: | Kuzin, A.P, Su, M, Vorobiev, S.M, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Northeast Structural Genomics Consortium Target XaR342
To be Published
|
|
4MHB
 
 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
4LTM
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4HRY
 
 | | The structure of Arabidopsis thaliana KAI2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Hydrolase, ... | | Authors: | Bythell-Douglas, R, Waters, M.T, Scaffidi, A, Flematti, G.R, Smith, S.M, Bond, C.S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Karrikin-Insensitive Protein (KAI2) in Arabidopsis thaliana
Plos One, 8, 2013
|
|
4HTA
 
 | | The structure of the karrikin insensitive (KAI2) protein in Arabidopsis thaliana | | Descriptor: | GLYCEROL, Hydrolase, alpha/beta fold family protein, ... | | Authors: | Bythell-Douglas, R, Waters, M.T, Scaffidi, A, Flematti, G.R, Smith, S.M, Bond, C.S. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The Structure of the Karrikin-Insensitive Protein (KAI2) in Arabidopsis thaliana
Plos One, 8, 2013
|
|
3S2Y
 
 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
1ZNP
 
 | | X-Ray Crystal Structure of Protein Q8U9W0 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR55. | | Descriptor: | hypothetical protein Atu3615 | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8U9W0 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium target AtR55.
To be Published
|
|
4AIF
 
 | | AIP TPR domain in complex with human Hsp90 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, HEAT SHOCK PROTEIN HSP 90-ALPHA, SULFATE ION | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
4APO
 
 | | AIP TPR domain in complex with human Tomm20 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, DODECAETHYLENE GLYCOL, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG, ... | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-04-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
4AEE
 
 | | CRYSTAL STRUCTURE OF MALTOGENIC AMYLASE FROM S.MARINUS | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Jung, T.Y, Park, C.H, Yoon, S.M, Park, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-18 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Association of Novel Domain in Active Site of Archaic Hyperthermophilic Maltogenic Amylase from Staphylothermus Marinus.
J.Biol.Chem., 287, 2012
|
|
1ZDR
 
 | | DHFR from Bacillus Stearothermophilus | | Descriptor: | GLYCEROL, SULFATE ION, dihydrofolate reductase | | Authors: | Kim, H.S, Damo, S.M, Lee, S.Y, Wemmer, D, Klinman, J.P. | | Deposit date: | 2005-04-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and hydride transfer mechanism of a moderate thermophilic dihydrofolate reductase from Bacillus stearothermophilus and comparison to its mesophilic and hyperthermophilic homologues.
Biochemistry, 44, 2005
|
|
4AQB
 
 | | MBL-Ficolin Associated Protein-1, MAP-1 aka MAP44 | | Descriptor: | CALCIUM ION, MANNAN-BINDING LECTIN SERINE PROTEASE 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skjoedt, M.O, Roversi, P, Hummelshoj, T, Palarasah, Y, Johnson, S, Lea, S.M, Garred, P. | | Deposit date: | 2012-04-16 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal Structure and Functional Characterization of the Complement Regulator Mannose-Binding Lectin (Mbl)/Ficolin-Associated Protein-1 (Map-1).
J.Biol.Chem., 287, 2012
|
|
6XFI
 
 | | Crystal Structures of beta-1,4-N-Acetylglucosaminyltransferase 2 (POMGNT2): Structural Basis for Inherited Muscular Dystrophies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ... | | Authors: | Halmo, S.M, Yeh, J, Wells, L, Moremen, K.W, Lanzilotta, W.N. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of beta-1,4-N-acetylglucosaminyltransferase 2: structural basis for inherited muscular dystrophies.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4AYI
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 Wild type | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, LIPOPROTEIN GNA1870 CCOMPND 7 | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
6XI2
 
 | | Apo form of POMGNT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-GLY-ALA-GLY-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Halmo, S.M, Yeh, J, Wells, L, Moremen, K.W, Lanzilotta, W.N. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structures of beta-1,4-N-acetylglucosaminyltransferase 2: structural basis for inherited muscular dystrophies.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4B0I
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) mutant (H70N) from Pseudomonas aeruginosa in complex with 3-hydroxydecanoyl-N-acetyl cysteamine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, S-[2-(acetylamino)ethyl] (3R)-3-hydroxydecanethioate | | Authors: | Moynie, L, McMahon, S.A, Leckie, S.M, Duthie, F.G, Koehnke, A, Naismith, J.H. | | Deposit date: | 2012-07-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonas Aeruginosa
J.Mol.Biol., 425, 2013
|
|
4B71
 
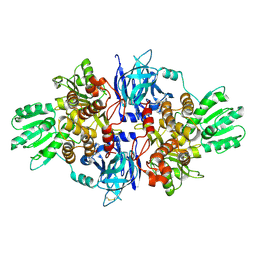 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propan-1-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4B7O
 
 | |
4AYN
 
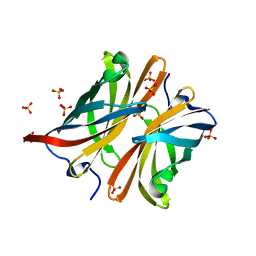 | | Structure of the C-terminal barrel of Neisseria meningitidis FHbp Variant 2 | | Descriptor: | FACTOR H-BINDING PROTEIN, SULFATE ION | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
6X61
 
 | |
4BKX
 
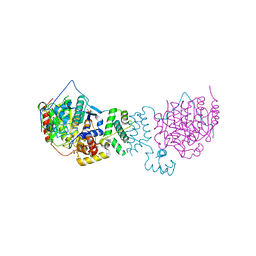 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
4BG9
 
 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (orthorhombic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL4
 
 | | Further structural insights into the binding of complement factor H by complement regulator acquiring surface protein 1, CspA (BbCRASP-1), of Borrelia burgdorferi. | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 1 (CRASP-1) | | Authors: | Caesar, J.J.E, Wallich, R, Kraiczy, P, Zipfel, P.F, Lea, S.M. | | Deposit date: | 2013-05-01 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.06 Å) | | Cite: | Further Structural Insights Into the Binding of Complement Factor H by Complement Regulator-Acquiring Surface Protein 1 (CspA) of Borrelia Burgdorferi.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
