1BQI
 
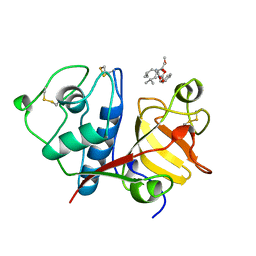 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | Descriptor: | CARBOBENZYLOXY-(L)-LEUCINYL-(L)LEUCINYL METHOXYMETHYLKETONE, PAPAIN | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
3N4E
 
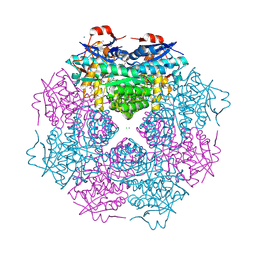 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans Pd1222 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans
Pd1222
To be Published
|
|
3N53
 
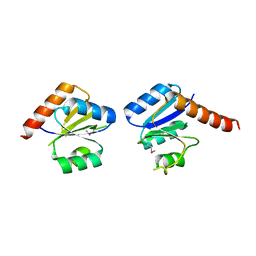 | |
2NWB
 
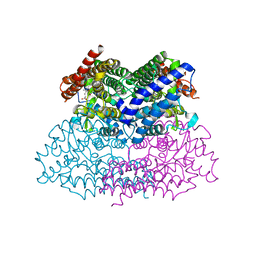 | | Crystal Structure of a Putative 2,3-dioxygenase (SO4414) from Shewanella oneidensis in complex with ferric heme. Northeast Structural Genomics Target SoR52. | | Descriptor: | Conserved domain protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Seetharaman, J, Bruckmann, C, Thackray, S.J, Khan, N, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Ma, L.C, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3N59
 
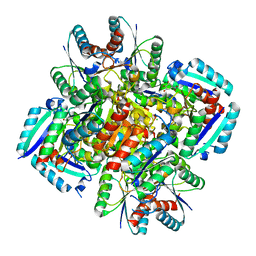 | | Type II dehydroquinase from Mycobacterium Tuberculosis complexed with 3-dehydroshikimate | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3OD3
 
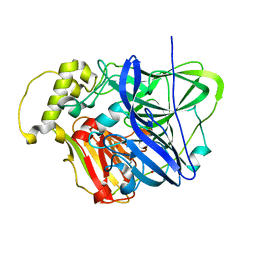 | | CueO at 1.1 A resolution including residues in previously disordered region | | Descriptor: | 1,2-ETHANEDIOL, Blue copper oxidase cueO, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2010-08-10 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3N1U
 
 | | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila | | Descriptor: | CALCIUM ION, Hydrolase, HAD superfamily, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila
To be published
|
|
3MY9
 
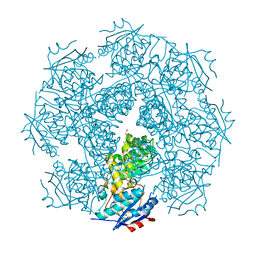 | | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Quartararo, C.E, Ramagopal, U, Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans
To be Published
|
|
3OF3
 
 | | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type 1 | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae
To be Published
|
|
3N07
 
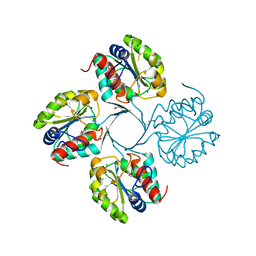 | | Structure of putative 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Vibrio cholerae | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, MAGNESIUM ION | | Authors: | Liu, W, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-04 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3OC4
 
 | | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase, pyridine nucleotide-disulfide family, ... | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583
To be Published
|
|
3N86
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 4 | | Descriptor: | (1R,5R)-1,5-dihydroxy-4-oxo-3-[3-oxo-3-(phenylamino)propyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3NFU
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 complexed with magnesium | | Descriptor: | GLYCEROL, Glucarate dehydratase, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3NHM
 
 | |
8T19
 
 | | RiPP precursor peptide recognition element (RRE) domain of Ocin-ThiF-like partner protein, PbtF, bound to an 8 residue fragment of its precursor peptide, PbtA | | Descriptor: | MAGNESIUM ION, PbtA, PbtF | | Authors: | Cogan, D.P, Nair, S.K, Mitchell, D.A. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Discovery and validation of RRE domains
To Be Published
|
|
3N76
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with compound 5 | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3Q8Y
 
 | |
3N2C
 
 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
2NW8
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and tryptophan. Northeast Structural Genomics Target XcR13. | | Descriptor: | MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3Q7M
 
 | |
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3N4F
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus sp. Y412MC10 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus
sp. Y412MC10
To be Published
|
|
3Q7N
 
 | |
3PAV
 
 | |
3OHP
 
 | |
