6KV3
 
 | |
6UKJ
 
 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
4WD9
 
 | |
7CA2
 
 | |
5VJ8
 
 | |
7RTY
 
 | |
6QZP
 
 | | High-resolution cryo-EM structure of the human 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S rRNA (1740-MER), 28S rRNA (3773-MER), ... | | Authors: | Natchiar, S.K, Myasnikov, A.G, Kratzat, H, Hazemann, I, Klaholz, B.P. | | Deposit date: | 2019-03-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of chemical modifications in the human 80S ribosome structure.
Nature, 551, 2017
|
|
2DVB
 
 | | Crystal structure of peanut lectin GAl-beta-1,6-GalNAc complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVF
 
 | | Crystals of peanut lectin grown in the presence of GAL-ALPHA-1,3-GAL-BETA-1,4-GAL | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVD
 
 | | Crystal structure of peanut lectin GAL-ALPHA-1,3-GAL complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DV9
 
 | | Crystal structure of peanut lectin GAL-BETA-1,3-GAL complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVA
 
 | | Crystal structure of peanut lectin GAL-BETA-1,3-GALNAC-ALPHA-O-ME (Methyl-T-antigen) complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVG
 
 | | Crystal structure of peanut lectin GAL-ALPHA-1,6-GLC complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
5G54
 
 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
2DH1
 
 | | Crystal structure of peanut lectin lactose-azobenzene-4,4'-dicarboxylic acid-lactose complex | | Descriptor: | Galactose-binding lectin | | Authors: | Natchiar, S.K, Srinivas, O, Nivedita, M, Sagarika, D, Jayaraman, N, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.65 Å) | | Cite: | Multivalency in lectins - A crystallographic, modelling and light-scattering study involving peanut lectin and a bivalent ligand
Curr.Sci., 90, 2006
|
|
2F6L
 
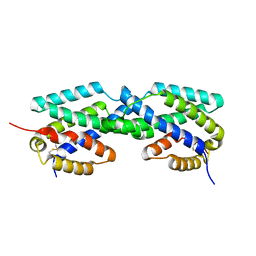 | | X-ray structure of Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | Chorismate mutase | | Authors: | Kim, S.K, Howard, A.J, Gilliland, G.L, Reddy, P.T, Ladner, J.E. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids.
J.Bacteriol., 188, 2006
|
|
1VTL
 
 | |
1VHY
 
 | |
1VOK
 
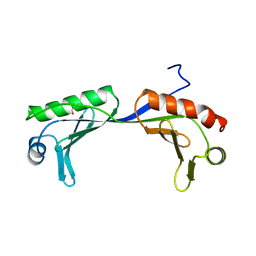 | | ARABIDOPSIS THALIANA TBP (DIMER) | | Descriptor: | SULFATE ION, TATA-BOX-BINDING PROTEIN | | Authors: | Nikolov, D.B, Burley, S.K. | | Deposit date: | 1996-04-29 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 A resolution refined structure of a TATA box-binding protein (TBP).
Nat.Struct.Biol., 1, 1994
|
|
8OYP
 
 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
1VQW
 
 | | Crystal structure of a protein with similarity to flavin-containing monooxygenases and to mammalian dimethylalanine monooxygenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN WITH SIMILARITY TO FLAVIN-CONTAINING MONOOXYGENASES AND TO MAMMALIAN DIMETHYLALANINE MONOOXYGENASES | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-05 | | Release date: | 2005-01-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1NKP
 
 | | Crystal structure of Myc-Max recognizing DNA | | Descriptor: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
8PXS
 
 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
