6XHM
 
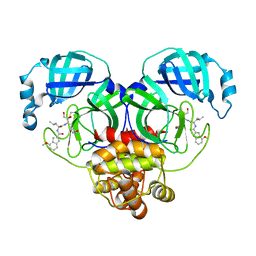 | | Covalent complex of SARS-CoV-2 main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHL
 
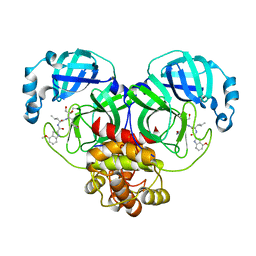 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHO
 
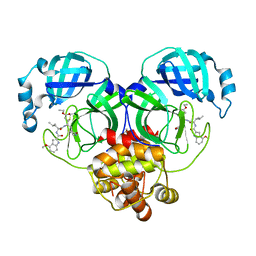 | | Covalent complex of SARS-CoV main protease with ethyl (4R)-4-({N-[(4-methoxy-1H-indol-2-yl)carbonyl]-L-leucyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, ethyl (2E,4S)-4-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
6XHN
 
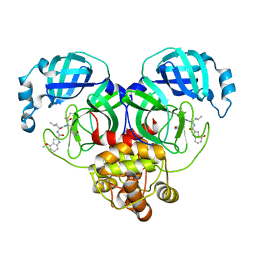 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Descriptor: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
1IA8
 
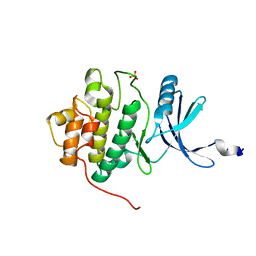 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
4I23
 
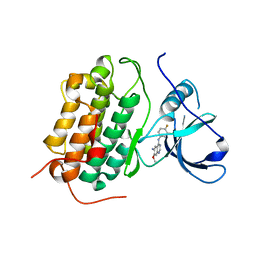 | | Crystal structure of the wild-type EGFR kinase domain in complex with dacomitinib (soaked) | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-methoxyquinazolin-6-yl}-4-(piperidin-1-yl)but-2-enamide, Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I21
 
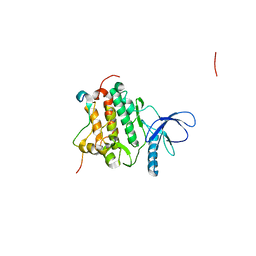 | | Crystal structure of L858R + T790M EGFR kinase domain in complex with MIG6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I20
 
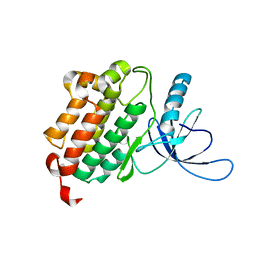 | | Crystal structure of monomeric (V948R) primary oncogenic mutant L858R EGFR kinase domain | | Descriptor: | Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I24
 
 | | Structure of T790M EGFR kinase domain co-crystallized with dacomitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-methoxyquinazolin-6-yl}-4-(piperidin-1-yl)but-2-enamide, Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I22
 
 | | Structure of the monomeric (V948R)gefitinib/erlotinib resistant double mutant (L858R+T790M) EGFR kinase domain co-crystallized with gefitinib | | Descriptor: | Epidermal growth factor receptor, Gefitinib, SULFATE ION | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
4I1Z
 
 | | Crystal structure of the monomeric (V948R) form of the gefitinib/erlotinib resistant EGFR kinase domain L858R+T790M | | Descriptor: | Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
2WGJ
 
 | | X-ray Structure of PF-02341066 bound to the kinase domain of c-Met | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Tran-Dube, M, Cui, J.J, Mroczkowski, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
2WKM
 
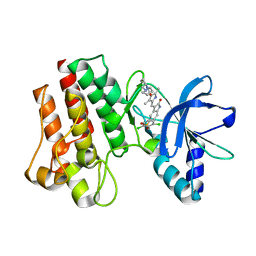 | | X-ray Structure of PHA-00665752 bound to the kinase domain of c-Met | | Descriptor: | (3Z)-5-[(2,6-DICHLOROBENZYL)SULFONYL]-3-[(3,5-DIMETHYL-4-{[(2S)-2-(PYRROLIDIN-1-YLMETHYL)PYRROLIDIN-1-YL]CARBONYL}-1H-PYRROL-2-YL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2009-06-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
7KK6
 
 | |
7KK4
 
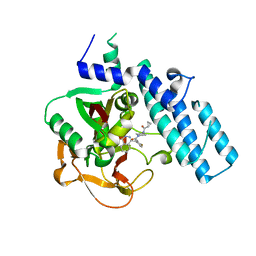 | | Structure of the catalytic domain of PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK3
 
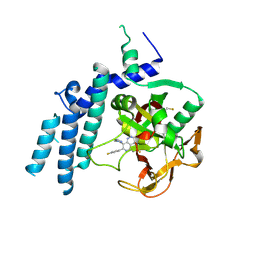 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKP
 
 | |
7KKM
 
 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKO
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKN
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Poly [ADP-ribose] polymerase, ... | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKQ
 
 | |
7KK2
 
 | | Structure of the catalytic domain of PARP1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK5
 
 | |
7LMC
 
 | |
3ZZE
 
 | | Crystal structure of C-MET kinase domain in complex with N'-((3Z)-4- chloro-7-methyl-2-oxo-1,2-dihydro-3H-indol-3-ylidene)-2-(4- hydroxyphenyl)propanohydrazide | | Descriptor: | (2S)-N'-[(3R)-4-chloro-7-methyl-2-oxo-2,3-dihydro-1H-indol-3-yl]-2-(4-hydroxyphenyl)propanehydrazide, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Deng, Y, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
