4PYN
 
 | | Humanized rat COMT in complex with SAH | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, POTASSIUM ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PYL
 
 | | Humanized rat COMT in complex with sinefungin, Mg2+, and tolcapone | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, SINEFUNGIN, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PYK
 
 | | human COMT, double domain swap | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PYM
 
 | | humanized rat apo-COMT bound to sulphate | | Descriptor: | Catechol O-methyltransferase, POTASSIUM ION, SULFATE ION | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PYI
 
 | | human apo COMT | | Descriptor: | Catechol O-methyltransferase, SODIUM ION | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5LQR
 
 | | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-(6-ethylpurin-9-yl)-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-(6-ethylpurin-9-yl)-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide
To be published
|
|
5LQN
 
 | |
5LQK
 
 | | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-[(4-methylphenyl)methyl]benzamide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of COMT
To be published
|
|
5LSA
 
 | |
3EAQ
 
 | |
3EAR
 
 | |
3EAS
 
 | |
2AFT
 
 | | Formylglycine generating enzyme C336S mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase modifying factor 1 | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-26 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AFY
 
 | | Formylglycine generating enzyme C341S mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase modifying factor 1 | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-26 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AII
 
 | | wild-type Formylglycine generating enzyme reacted with iodoacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, CALCIUM ION, ... | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3TRK
 
 | | Structure of the Chikungunya virus nsP2 protease | | Descriptor: | Nonstructural polyprotein, SODIUM ION | | Authors: | Cheung, J, Franklin, M, Mancia, F, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure of the Chikungunya virus nsP2 protease
To be Published
|
|
2AIK
 
 | |
2AIJ
 
 | |
3NWB
 
 | | Rat COMT in complex with a fluorinated desoxyribose-containing bisubstrate inhibitor avoids hydroxyl group | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-07-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3R6T
 
 | | Rat catechol o-methyltransferase in complex with the bisubstrate inhibitor 4'-fluoro-4,5-dihydroxy-biphenyl-3-carboxylic acid {(E)-3-[(2S,4R,5R)-4-hydroxy-5-(6-methyl-purin-9-yl)-tetrahydro-furan-2-yl]-allyl}-amide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4'-fluoro-4,5-dihydroxy-N-{(2E)-3-[(2S,4R,5R)-4-hydroxy-5-(6-methyl-9H-purin-9-yl)tetrahydrofuran-2-yl]prop-2-en-1-yl}biphenyl-3-carboxamide, CHLORIDE ION, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3S68
 
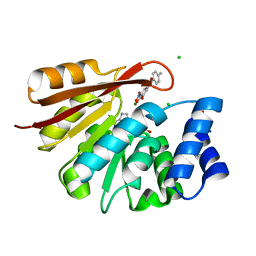 | | Rat COMT in complex with SAM and Tolcapone at 1.85A, P3221, Rfree=22.0 | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2011-05-25 | | Release date: | 2012-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OE4
 
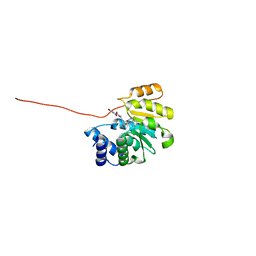 | | Rat catechol O-methyltransferase in complex with a catechol-type, purine-containing bisubstrate inhibitor - humanized form | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, N-[(E)-3-[(2R,3S,4R,5R)-3,4-dihydroxy-5-purin-9-yl-oxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-nitro-benzamide | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
3NW9
 
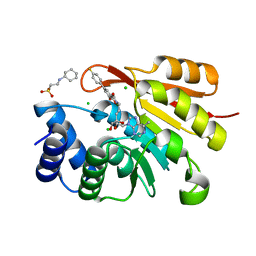 | | Rat COMT in complex with a methylpurin-containing bisubstrate inhibitor | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-07-09 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
3OE5
 
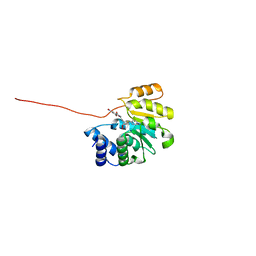 | | Rat catechol O-methyltransferase in complex with a catechol-type, pyridylsulfanyl-containing inhibitor - humanized form | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, N-[(E)-3-[(2R,3S,4R,5S)-3,4-dihydroxy-5-pyridin-4-ylsulfanyl-oxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-nitro-benzamide | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
3OZT
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, 4-oxo-pyridinyl-containing inhibitor - humanized form | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, N-[(E)-3-[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-oxopyridin-1-yl)oxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-nitro-benzamide | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
