1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | Descriptor: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
1T0N
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
7SKY
 
 | | Pertussis toxin S1 bound to NAD+ | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37000132 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKI
 
 | | Pertussis toxin in complex with PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.09912443 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKK
 
 | | pertussis toxin in complex with ADPR and Nicotinamide | | Descriptor: | NICOTINAMIDE, Pertussis toxin subunit 1, SULFATE ION, ... | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65000772 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7T6I
 
 | | Crystal structure of HLA-DP1 in complex with pp65 peptide in reverse orientation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Lim, J.J, Reid, H, Rossjohn, J. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human T cells recognize HLA-DP-bound peptides in two orientations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U6Z
 
 | | Pertussis toxin E129D NAD | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.30002 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
6MSS
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | (2S)-2-(heptadecanoyloxy)-3-{[(10S)-10-methyloctadecanoyl]oxy}propyl alpha-D-glucopyranosiduronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
7JWI
 
 | |
7JWJ
 
 | | Crystal Structure of B17-C1 TCR-H2Db | | Descriptor: | B17.C1 TCR alpha chain, B17.C1 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Farenc, C, Rossjohn, J, Gras, S, Szeto, C. | | Deposit date: | 2020-08-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling.
Science, 372, 2021
|
|
6MRA
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | TCR alpha chain, TCR beta-chain | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
4P15
 
 | | Structure of the ClpC N-terminal domain from an alkaliphilic Bacillus lehensis G1 species | | Descriptor: | Bacillus lehensis ClpC N-terminal domain, SULFATE ION | | Authors: | Rashid, S.A, Littler, D.R, Illias, R.M, Murad, A.M.A, Rossjohn, J, Beddoe, T, Mahadi, N.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the ClpC N-terminal domain at 1.85 Angstroms resolution from an alkaliphilic Bacillus lehensis G1 species
To Be Published
|
|
6NUX
 
 | | CD1a-lipid binary complex | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2019-02-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human T cell response to CD1a and contact dermatitis allergens in botanical extracts and commercial skin care products.
Sci Immunol, 5, 2020
|
|
8UMO
 
 | | Murine CD94-NKG2A receptor in complex with Qa-1b presenting AMAPRTLLL | | Descriptor: | BROMIDE ION, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2023-10-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the murine CD94-NKG2A receptor in complex with Qa-1 b presenting an MHC-I leader peptide.
Febs J., 291, 2024
|
|
7T0L
 
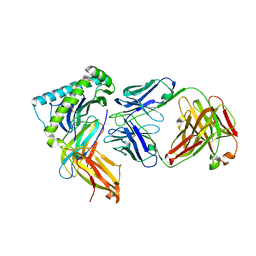 | |
8VZ8
 
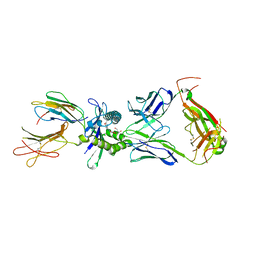 | | Crystal structure of mouse MAIT M2B TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
8VZ9
 
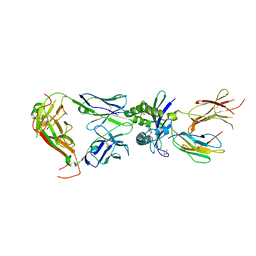 | | Crystal structure of mouse MAIT M2A TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
7JPT
 
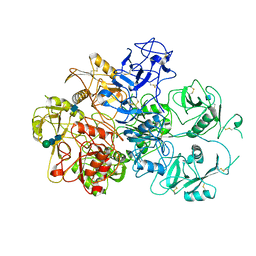 | | Structure of an endocytic receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 75, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gully, B.S, Rossjohn, J, Berry, R. | | Deposit date: | 2020-08-09 | | Release date: | 2020-12-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of the endocytic receptor DEC-205.
J.Biol.Chem., 296, 2020
|
|
7K0Z
 
 | | Marsupial T cell receptor Spl_157 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, T cell receptor mu chain | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-09-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
7K0X
 
 | | Marsupial T cell receptor Spl_145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, T cell receptor mu chain, ... | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-09-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
1PY9
 
 | | The crystal structure of an autoantigen in multiple sclerosis | | Descriptor: | Myelin-oligodendrocyte glycoprotein, SULFATE ION | | Authors: | Clements, C.S, Reid, H.H, Beddoe, T, Tynan, F.E, Perugini, M.A, Johns, T.G, Bernard, C.C, Rossjohn, J. | | Deposit date: | 2003-07-08 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7JYV
 
 | | Crystal Structure of HLA A*2402 in complex with YFSPIRVTF, an 9-mer influenza epitope | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7JYU
 
 | | Crystal Structure of HLA-A*2402 in complex with IYFSPIRVTF, an 10-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Nguyen, A.T, Szeto, C, Rossjohn, J, Gras, S. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7JYW
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWIIRNW, an 9-mer influenza epitope | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, MHC class I antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7JYX
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWIIRNWET, an 11-mer epitope from Influenza | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PB2 peptide from Influenza, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
