7PVI
 
 | | dTDP-sugar epimerase | | Descriptor: | CITRATE ANION, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWB
 
 | | dTDP-sugar epimerase from Coxiella burnetii in complex with dTDP | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWI
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, dTDP-4-keto-rhamnose 3,5-epimerase,dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.326 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWH
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
1GN0
 
 | | Escherichia coli GlpE sulfurtransferase soaked with KCN | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-10-01 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
1GMX
 
 | | Escherichia coli GlpE sulfurtransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
1URH
 
 | | The "Rhodanese" fold and catalytic mechanism of 3-mercaptopyruvate sulfotransferases: Crystal structure of SseA from Escherichia coli | | Descriptor: | 3-MERCAPTOPYRUVATE SULFURTRANSFERASE, SULFITE ION | | Authors: | Spallarossa, A, Forlani, F, Carpen, A, Armirotti, A, Pagani, S, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-10-30 | | Release date: | 2003-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The "Rhodanese" Fold and Catalytic Mechanism of 3-Mercaptopyruvate Sulfurtransferases: Crystal Structure of Ssea from Escherichia Coli
J.Mol.Biol., 335, 2004
|
|
2VG7
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-iodophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG6
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG5
 
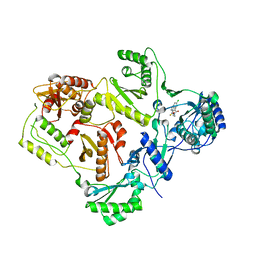 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
5IO9
 
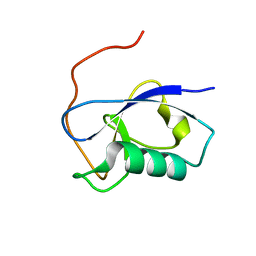 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IP4
 
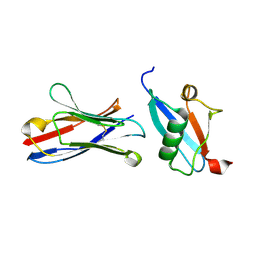 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IN7
 
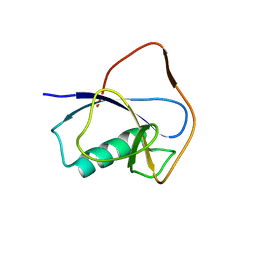 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IOI
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
1GNC
 
 | |
1OKD
 
 | | NMR-structure of tryparedoxin 1 | | Descriptor: | TRYPAREDOXIN 1 | | Authors: | Krumme, D, Budde, H, Hecht, H.-J, Menge, U, Ohlenschlager, O, Ross, A, Wissing, J, Wray, V, Flohe, L. | | Deposit date: | 2003-07-22 | | Release date: | 2003-08-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR studies of the interaction of tryparedoxin with redox-inactive substrate homologues.
Biochemistry, 42, 2003
|
|
1JAS
 
 | | HsUbc2b | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA | | Authors: | Miura, T, Klaus, W, Ross, A, Guentert, P, Senn, H. | | Deposit date: | 2001-05-31 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the class I human ubiquitin-conjugating enzyme 2b
J.Biomol.NMR, 22, 2002
|
|
1JXN
 
 | | Crystal Structure of the Lectin I from Ulex europaeus in complex with the methyl glycoside of alpha-L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Audette, G.F, Olson, D.J.H, Ross, A.R.S, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2001-09-07 | | Release date: | 2002-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Examination of the Structural Basis for O(H) Blood Group Specificity by Ulex europaeus Lectin I
Can.J.Chem., 80, 2002
|
|
7OVY
 
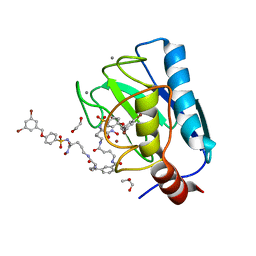 | | Crystal structure of a dimeric based inhibitor JG34 in complex with the MMP-12 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Ciccone, L, Rossello, A, Vera, L, Nuti, E, Stura, E.A. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of Dimeric Arylsulfonamides as Potent ADAM8 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6TI9
 
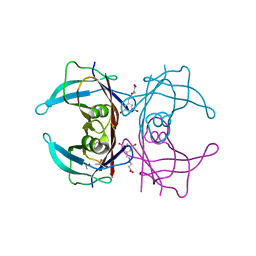 | | Human transthyretin (TTR) complexed with (E)-3-(((3,5-dibromo-2-hydroxybenzylidene)amino)oxy)propanoic acid. | | Descriptor: | 3-[(~{E})-[3,5-bis(bromanyl)-2-oxidanyl-phenyl]methylideneamino]oxypropanoic acid, GLYCEROL, S-1,2-PROPANEDIOL, ... | | Authors: | Ciccone, L, Nencetti, S, Orlandini, E, Rossello, A, Legrand, P, Shepard, W. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Monoaryl derivatives as transthyretin fibril formation inhibitors: Design, synthesis, biological evaluation and structural analysis.
Bioorg.Med.Chem., 28, 2020
|
|
6TJN
 
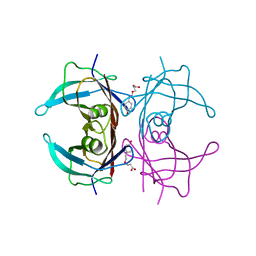 | | Human transthyretin (TTR) complexed with (E)-3-(((4-hydroxybenzylidene)amino)oxy)propanoic acid | | Descriptor: | 3-[(~{E})-(4-hydroxyphenyl)methylideneamino]oxypropanoic acid, Transthyretin | | Authors: | Ciccone, L, Shepard, W, Nencetti, S, Orlandini, E, Rossello, A. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Monoaryl derivatives as transthyretin fibril formation inhibitors: Design, synthesis, biological evaluation and structural analysis.
Bioorg.Med.Chem., 28, 2020
|
|
9C50
 
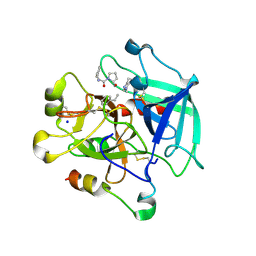 | | Replacement of a single residue changes the primary specificity of thrombin | | Descriptor: | FPF, SODIUM ION, Thrombin A-chain, ... | | Authors: | Dei Rossi, A, Deavila, S, Mohammed, B.M, Korolev, S, Di Cera, E. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Replacement of a single residue changes the primary specificity of thrombin.
J.Thromb.Haemost., 23, 2025
|
|
7Q4I
 
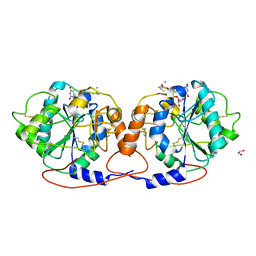 | | Crystal structure of DmC1GalT1 in complex with UDP-Mn2+ and the APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1, ... | | Authors: | Gonzalez-Ramirez, A.M, Coelho, H, Companon, I, Grosso, A.S, Yang, Z, Narimatsu, Y, Clausen, H, Marcelo, F, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2021-10-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the synthesis of the core 1 structure by C1GalT1.
Nat Commun, 13, 2022
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
4TQH
 
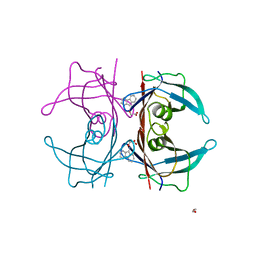 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)ethanoic acid | | Descriptor: | 1,2-ETHANEDIOL, Transthyretin, [(9H-fluoren-9-ylideneamino)oxy]acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
