3FEY
 
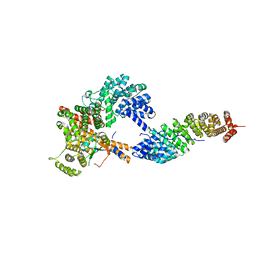 | |
3EWK
 
 | |
3FBV
 
 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
4H49
 
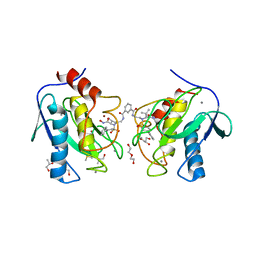 | | Crystal structure of the catalytic domain of MMP-12 in complex with a twin inhibitor. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Antoni, C, Stura, E.A, Vera, L, Nuti, E, Carafa, L, Cassar-Lajeunesse, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4H1Q
 
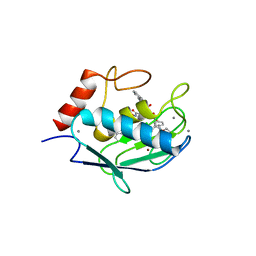 | | Crystal structure of mutant MMP-9 catalytic domain in complex with a twin inhibitor. | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, N-(4-{[(3R)-3-[(biphenyl-4-ylsulfonyl)(propan-2-yloxy)amino]-4-(hydroxyamino)-4-oxobutyl]amino}-4-oxobutyl)-N'-(4-{[(3S)-3-[(biphenyl-4-ylsulfonyl)(propan-2-yloxy)amino]-4-(hydroxyamino)-4-oxobutyl]amino}-4-oxobutyl)benzene-1,3-dicarboxamide, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Catalani, M.P, Dive, V, Rossello, A. | | Deposit date: | 2012-09-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4M1F
 
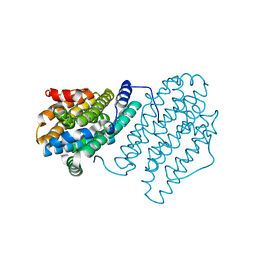 | |
3FDC
 
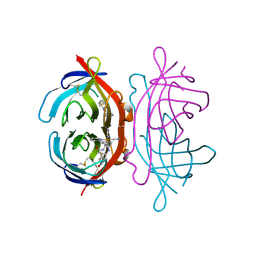 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
1ZYE
 
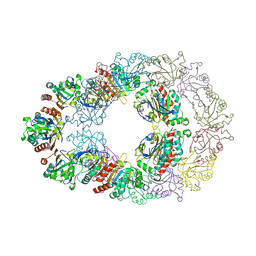 | | Crystal structure analysis of Bovine Mitochondrial Peroxiredoxin III | | Descriptor: | Thioredoxin-dependent peroxide reductase | | Authors: | Cao, Z, Roszak, A.W, Gourlay, L.J, Lindsay, J.G, Isaacs, N.W. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bovine Mitochondrial Peroxiredoxin III Forms a Two-Ring Catenane
Structure, 13, 2005
|
|
3FEX
 
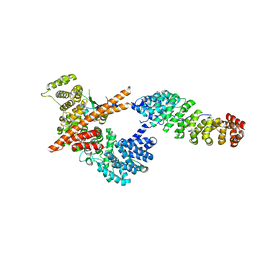 | |
1YEW
 
 | |
4FXM
 
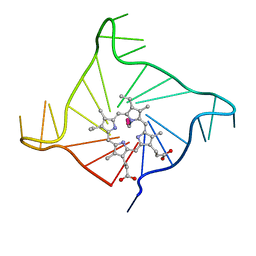 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
4H2E
 
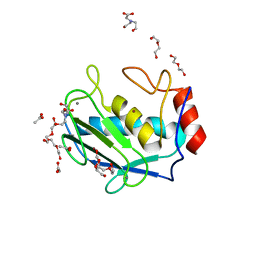 | | Crystal structure of an MMP twin inhibitor complexing two MMP-9 catalytic domains | | Descriptor: | ACETATE ION, BICINE, CALCIUM ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Catalani, M.P, Dive, V, Rossello, A. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
2ABO
 
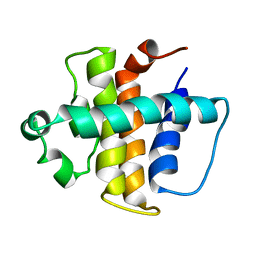 | | NMR structure of gamma herpesvirus 68 a viral Bcl-2 homolog | | Descriptor: | bcl-2 homolog | | Authors: | Loh, J, Huang, Q, Petros, A.M, Nettesheim, D, van Dyk, L.F, Labrada, L, Speck, S.H, Levine, B, Olejniczak, E.T, Virgin, H.W. | | Deposit date: | 2005-07-15 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A surface groove essential for viral Bcl-2 function during chronic infection in vivo.
Plos Pathog., 1, 2005
|
|
4H84
 
 | | Crystal structure of the catalytic domain of Human MMP12 in complex with a selective carboxylate based inhibitor. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Antoni, C, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-21 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4KRY
 
 | | Structure of Aes from E. coli in covalent complex with PMS | | Descriptor: | Acetyl esterase, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
3G5W
 
 | | Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea | | Descriptor: | COPPER (II) ION, CU-O LINKAGE, CU-O-CU LINKAGE, ... | | Authors: | Lawton, T.J, Sayavedra-Soto, L.A, Arp, D.J, Rosenzweig, A.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a two-domain multicopper oxidase: implications for the evolution of multicopper blue proteins.
J.Biol.Chem., 284, 2009
|
|
3FRY
 
 | | Crystal structure of the CopA C-terminal metal binding domain | | Descriptor: | CITRIC ACID, Probable copper-exporting P-type ATPase A | | Authors: | Agarwal, S, Sazinsky, M, Arguello, J, Rosenzweig, A.C. | | Deposit date: | 2009-01-08 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA.
Proteins, 78, 2010
|
|
4M1H
 
 | | X-ray crystal structure of Chlamydia trachomatis apo NrdB | | Descriptor: | Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Basis for Assembly of the Mn(IV)/Fe(III) Cofactor in the Class Ic Ribonucleotide Reductase from Chlamydia trachomatis.
Biochemistry, 52, 2013
|
|
4N59
 
 | | The Crystal Structure of Pectocin M2 at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Pectocin M2, ... | | Authors: | Zeth, K, Grinter, R, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4JJM
 
 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
4UUJ
 
 | | POTASSIUM CHANNEL KCSA-FAB WITH TETRAHEXYLAMMONIUM | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Burdette, D, Wagner, T, Focia, P.J, Gross, A. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
2XYF
 
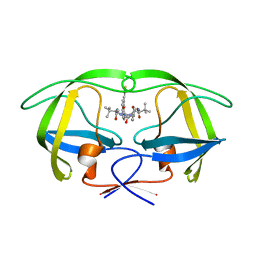 | | HIV-1 Inhibitors with a Tertiary-Alcohol-containing Transition-State Mimic and various P2 and P1 prime Substituents | | Descriptor: | METHYL N-[(2S)-1-[2-[(4R)-5-[[(2S)-3,3-DIMETHYL-1-METHYLAMINO-1-OXO-BUTAN-2-YL]AMINO]-4-HYDROXY-5-OXO-4-(PHENYLMETHYL)PENTYL]-2-[(4-THIOPHEN-3-YLPHENYL)METHYL]HYDRAZINYL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, PROTEASE | | Authors: | Ohrngren, P, Wu, X, Persson, M, Ekegren, J.K, Wallberg, H, Rosenquist, A, Samuelsson, B, Unge, T, Larhed, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 Protease Inhibitors with a Tertiary Alcohol Containing Transition-State Mimic and Various P2 and P1' Substituents
Med.Chem.Commun., 2, 2011
|
|
2Y2K
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
2XYE
 
 | | HIV-1 Inhibitors with a Tertiary-Alcohol-containing Transition-State Mimic and various P2 and P1 prime Substituents | | Descriptor: | METHYL N-[(2S)-1-[2-[(4R)-5-[[(2S)-3,3-DIMETHYL-1-METHYLAMINO-1-OXO-BUTAN-2-YL]AMINO]-4-HYDROXY-5-OXO-4-(PHENYLMETHYL)PENTYL]-2-[(4-PHENYLPHENYL)METHYL]HYDRAZINYL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, PROTEASE | | Authors: | Ohrngren, P, Wu, X, Persson, M, Ekegren, J.K, Wallberg, H, Rosenquist, A, Samuelsson, B, Unge, T, Larhed, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors with a Tertiary Alcohol Containing Transition-State Mimic and Various P2 and P1' Substituents
Med.Chem.Commun., 2, 2011
|
|
3J6Y
 
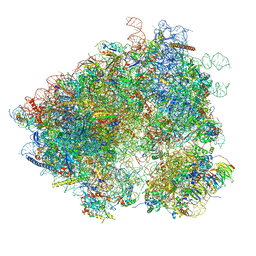 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 2 degree rotation (Class I) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
