4V7P
 
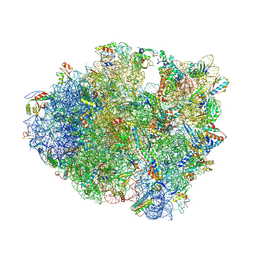 | | Recognition of the amber stop codon by release factor RF1. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Zhu, J, Asahara, H, Noller, H.F. | | Deposit date: | 2010-04-29 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition of the amber UAG stop codon by release factor RF1.
Embo J., 29, 2010
|
|
4V4I
 
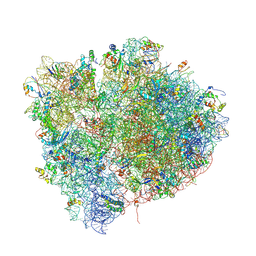 | | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements. | | Descriptor: | 16S SMALL SUBUNIT RIBOSOMAL RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-02-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements
Cell(Cambridge,Mass.), 126, 2006
|
|
1W7B
 
 | |
8EC6
 
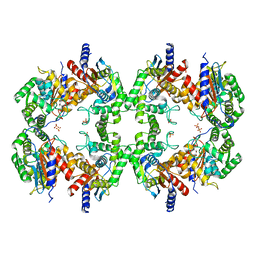 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PVI
 
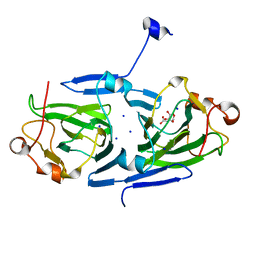 | | dTDP-sugar epimerase | | Descriptor: | CITRATE ANION, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWB
 
 | | dTDP-sugar epimerase from Coxiella burnetii in complex with dTDP | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWI
 
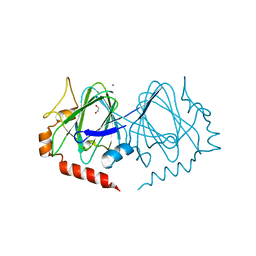 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, dTDP-4-keto-rhamnose 3,5-epimerase,dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.326 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWH
 
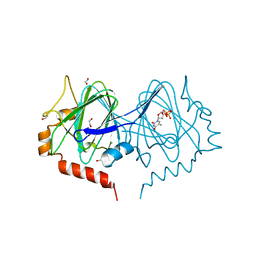 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
6TIK
 
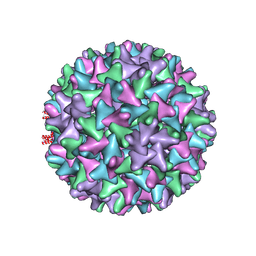 | | Hepatitis B virus core shell--virus-like particle with NadA epitope | | Descriptor: | Capsid protein,Putative adhesin/invasin,Capsid protein,Factor H-binding protein | | Authors: | Roseman, A.M, Colllins, R.F, Derrick, J.P. | | Deposit date: | 2019-11-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An assessment of the use of Hepatitis B Virus core protein virus-like particles to display heterologous antigens from Neisseria meningitidis.
Vaccine, 38, 2020
|
|
7ZBU
 
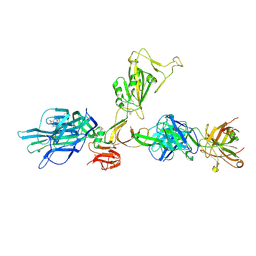 | | CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, P008_60 antibody, ... | | Authors: | Rosa, A, Pye, V.E, Cronin, N, Cherepanov, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Cell Rep, 40, 2022
|
|
7NT9
 
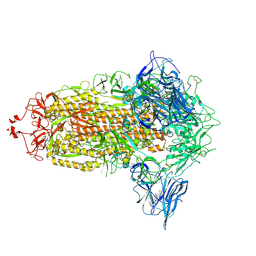 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTA
 
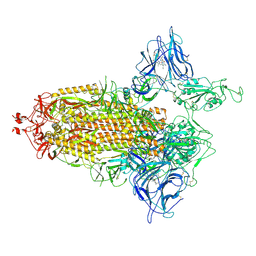 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTC
 
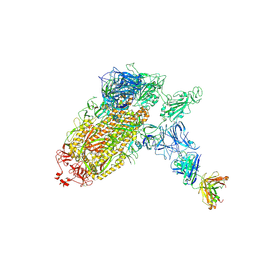 | | Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
6VKL
 
 | | Negative stain reconstruction of the yeast exocyst octameric complex. | | Descriptor: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | Authors: | Frost, A, Munson, M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Exocyst structural changes associated with activation of tethering downstream of Rho/Cdc42 GTPases.
J. Cell Biol., 219, 2020
|
|
1MMO
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL NON-HAEM IRON HYDROXYLASE THAT CATALYSES THE BIOLOGICAL OXIDATION OF METHANE | | Descriptor: | ACETIC ACID, FE (III) ION, METHANE MONOOXYGENASE HYDROLASE (ALPHA CHAIN), ... | | Authors: | Rosenzweig, A.C, Frederick, C.A, Lippard, S.J, Nordlund, P. | | Deposit date: | 1994-02-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane.
Nature, 366, 1993
|
|
2HG3
 
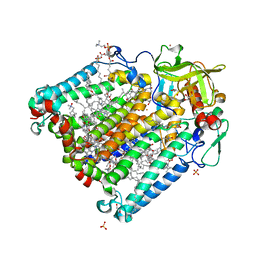 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with brominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HH1
 
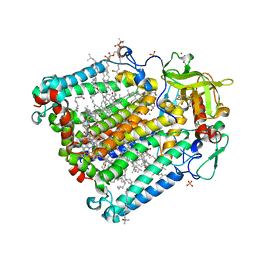 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
1P0C
 
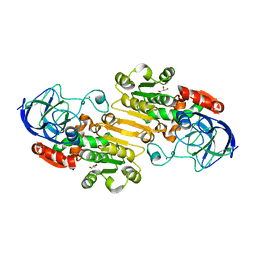 | | Crystal Structure of the NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) | | Descriptor: | GLYCEROL, NADP-dependent ALCOHOL DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Vertebrate NADP(H)-dependent Alcohol Dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
2HIT
 
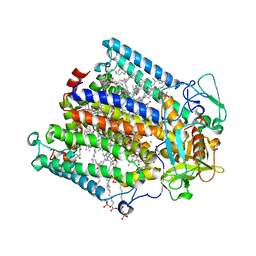 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylethanolamine | | Descriptor: | (1R)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HHK
 
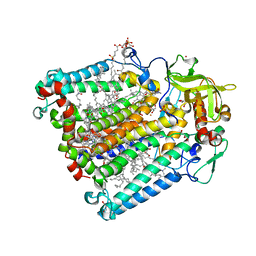 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylglycerol | | Descriptor: | (1R)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HG9
 
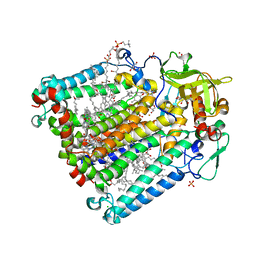 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with tetrabrominated phosphatidylcholine | | Descriptor: | (7R,18S,19R)-18,19-DIBROMO-7-{[(9S,10S)-9,10-DIBROMOOCTADECANOYL]OXY}-4-HYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-P HOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
1CC8
 
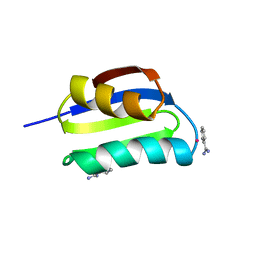 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, MERCURY (II) ION, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
5U5P
 
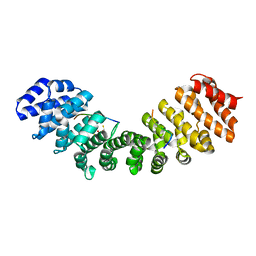 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus Musculus Complexed with a MLH1 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
1G5J
 
 | | COMPLEX OF BCL-XL WITH PEPTIDE FROM BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BAD PROTEIN | | Authors: | Petros, A.M, Nettesheim, D.G, Wang, Y, Olejniczak, E.T, Meadows, R.P, Mack, J, Swift, K, Matayoshi, E.D, Zhang, H, Thompson, C.B, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies.
Protein Sci., 9, 2000
|
|
1GN0
 
 | | Escherichia coli GlpE sulfurtransferase soaked with KCN | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-10-01 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
