1E4E
 
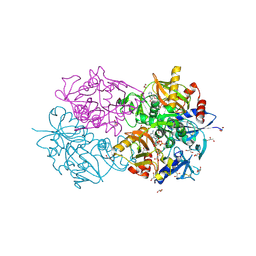 | | D-alanyl-D-lacate ligase | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roper, D.I. | | Deposit date: | 2000-07-03 | | Release date: | 2001-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA).
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
1GQZ
 
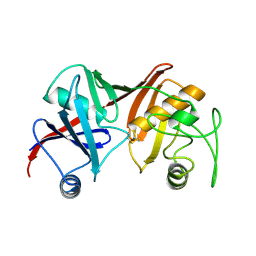 | |
7ZG8
 
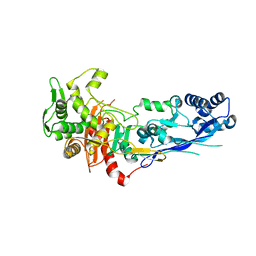 | |
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
5UG1
 
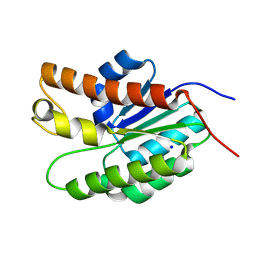 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain with methylsulfonyl adduct | | Descriptor: | Acyltransferase, SODIUM ION, methanesulfonic acid | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UFY
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acyltransferase, SODIUM ION | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
6R1N
 
 | | Crystal structure of S. aureus seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
6R1O
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to a seryl sulfamoyl adenosine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
6R1M
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, PHOSPHATE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
6FTB
 
 | | Staphylococcus aureus monofunctional glycosyltransferase in complex with moenomycin | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, 1,2-ETHANEDIOL, MOENOMYCIN, ... | | Authors: | Punekar, A.S, Dowson, C.J, Roper, D.I. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of the jaw subdomain of peptidoglycan glycosyltransferases for lipid II polymerization.
Cell Surf, 2, 2018
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
3FVE
 
 | | Crystal structure of diaminopimelate epimerase Mycobacterium tuberculosis DapF | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Diaminopimelate epimerase, GLYCEROL | | Authors: | Usha, V, Dover, L.G, Roper, D.I, Futterer, K, Besra, G.S. | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2OTN
 
 | | Crystal structure of the catalytically active form of diaminopimelate epimerase from Bacillus anthracis | | Descriptor: | Diaminopimelate epimerase | | Authors: | Matho, M.H, Fukuda, K, Santelli, E, Jaroszewski, L, Liddington, R.C, Roper, D. | | Deposit date: | 2007-02-08 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and inhibition of a catalytically active form of diaminopimelate epimerase (DapF)from Bacillus anthracis
To be Published
|
|
4C12
 
 | | X-ray Crystal Structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fulop, V, Roper, D.I, Ruane, K.M, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity Determinants for Lysine Incorporation in Staphylococcus Aureus Peptidoglycan as Revealed by the Structure of a Mure Enzyme Ternary Complex.
J.Biol.Chem., 288, 2013
|
|
4C5C
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ADP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5A
 
 | | The X-ray crystal structures of D-alanyl-D-alanine ligase in complex ADP and D-cycloserine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, GLYCEROL, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5B
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBONATE ION, D-ALANINE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
3A3F
 
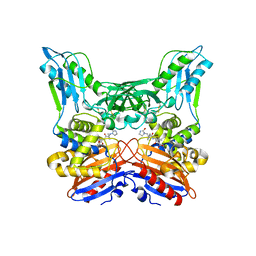 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae,complexed with novel beta-lactam (FMZ) | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-({(2R)-2-[(2-oxoimidazolidin-1-yl)amino]-2-phenylacetyl}amino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3I
 
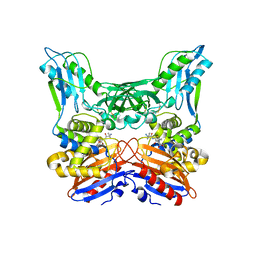 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with ampicillin (AIX) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3J
 
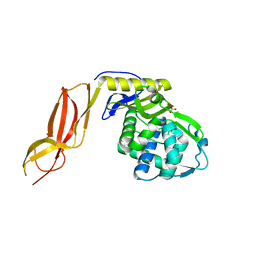 | | Crystal structures of penicillin binding protein 5 from Haemophilus influenzae | | Descriptor: | PBP5, SULFATE ION | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3D
 
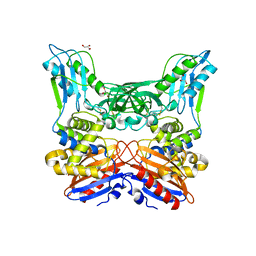 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae | | Descriptor: | GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
2VWT
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
2VWS
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 | | Descriptor: | GLYCEROL, PHOSPHATE ION, YFAU, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
3A3E
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with novel beta-lactam (CMV) | | Descriptor: | (2R,4S)-2-[(1R)-1-({(2R)-2-[(4-ethyl-2,3-dioxopiperazin-1-yl)amino]-2-phenylacetyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
