1YA7
 
 | | Implications for interactions of proteasome with PAN and PA700 from the 1.9 A structure of a proteasome-11S activator complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAU
 
 | | Structure of Archeabacterial 20S proteasome- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAR
 
 | | Structure of Archeabacterial 20S proteasome mutant D9S- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
4OJV
 
 | | Crystal structure of unliganded yeast PDE1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, SULFATE ION, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
4OJX
 
 | | crystal structure of yeast phosphodiesterase-1 in complex with GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
2R5C
 
 | | Aedes Kynurenine Aminotransferase in Complex with Cysteine | | Descriptor: | Kynurenine aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
1ZNO
 
 | | Crystal Structure of VC702 from Vibrio Cholerae, Northeast Structural Genomics Consortium Target: VcP1 | | Descriptor: | Hypothetical UPF0244 protein VC0702, MAGNESIUM ION | | Authors: | Ni, S, Forouhar, F, Bussiere, D.E, Robinson, H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2006-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of VC0702 at 2.0 A: conserved hypothetical protein from Vibrio cholerae.
Proteins, 63, 2006
|
|
1TYZ
 
 | | Crystal structure of 1-Aminocyclopropane-1-carboyxlate Deaminase from Pseudomonas | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes:Insight into the mechanism of unique pyrodoxial-5'-phosphate dependent cyclopropane ring opening reaction
Biochemistry, 43, 2004
|
|
1TZK
 
 | | Crystal structure of 1-aminocyclopropane-1-carboxylate-deaminase complexed with alpha-keto-butyrate | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 2-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
3S2Y
 
 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
212D
 
 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
220D
 
 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*AP*CP*CP*CP*GP*CP*GP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
221D
 
 | |
222D
 
 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | COBALT HEXAMMINE(III), DNA/RNA (5'-R(*GP*CP*)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
4QLW
 
 | | Azurin mutant M121E with iron | | Descriptor: | Azurin, FE (III) ION, NITRATE ION, ... | | Authors: | Liu, J, Robinson, H, Lu, Y. | | Deposit date: | 2014-06-13 | | Release date: | 2014-08-13 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redesigning the Blue Copper Azurin into a Redox-Active Mononuclear Nonheme Iron Protein: Preparation and Study of Fe(II)-M121E Azurin.
J.Am.Chem.Soc., 136, 2014
|
|
4QKT
 
 | | Azurin mutant M121EM44K with copper | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Azurin, ... | | Authors: | Liu, J, Robinson, H, Lu, Y. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Redesigning the Blue Copper Azurin into a Redox-Active Mononuclear Nonheme Iron Protein: Preparation and Study of Fe(II)-M121E Azurin.
J.Am.Chem.Soc., 136, 2014
|
|
3E4B
 
 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | Descriptor: | AlgK, CHLORIDE ION, GLYCEROL | | Authors: | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
6BDC
 
 | |
4HU8
 
 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
3V52
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, ... | | Authors: | Mage, M.G, Dolan, M.A, Wang, R, Boyd, L.F, Revilleza, M.J, Robinson, H, Natarajan, K, Myers, N.B, Hansen, T.H, Margulies, D.H. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3T6A
 
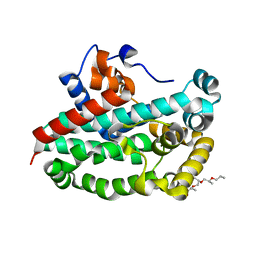 | | Structure of the C-terminal domain of BCAR3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, Breast cancer anti-estrogen resistance protein 3, UNKNOWN ATOM OR ION | | Authors: | Mace, P.D, Robinson, H, Riedl, S.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NSP-Cas protein structures reveal a promiscuous interaction module in cell signaling.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3T6G
 
 | |
3M38
 
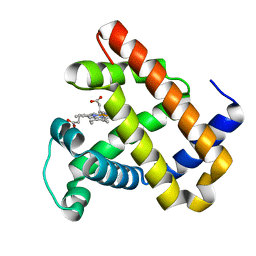 | | The roles of Glutamates and Metal ions in a rationally designed nitric oxide reductase based on myoglobin: I107E FeBMb (No metal ion binding to FeB site) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TS5
 
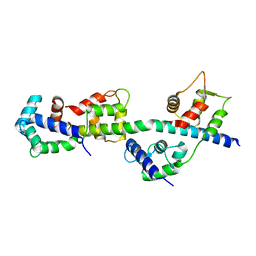 | | Crystal Structure of a Light Chain Domain of Scallop Smooth Muscle Myosin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Kumar, V.S.S, O'Neall-Hennessey, E, Reshetnikova, L, Brown, J.H, Robinson, H, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2011-09-12 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle Myosin.
Biophys.J., 101, 2011
|
|
5EQ6
 
 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
