4A5U
 
 | |
1HSN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
2ARV
 
 | | Structure of human Activin A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Inhibin beta A chain, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
3K7R
 
 | | Crystal structure of [TM][CuAtx1]3 | | Descriptor: | COPPER (II) ION, D-MALATE, Metal homeostasis factor ATX1, ... | | Authors: | Xue, Y, Alvarez, H.M, Robinson, C.D, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation.
Science, 327, 2010
|
|
1GQ6
 
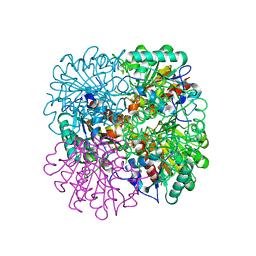 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric Structure of Proclavaminic Acid Amidino Hydrolase: Evolution of a Hydrolytic Enzyme in Clavulanic Acid Biosynthesis
Biochem.J., 366, 2002
|
|
1GQ7
 
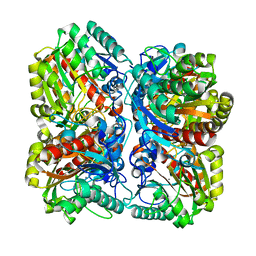 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Oligomeric structure of proclavaminic acid amidino hydrolase: evolution of a hydrolytic enzyme in clavulanic acid biosynthesis.
Biochem. J., 366, 2002
|
|
1I11
 
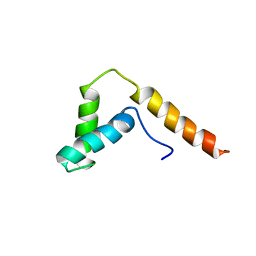 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN, SOX-5 HMG BOX FROM MOUSE | | Descriptor: | TRANSCRIPTION FACTOR SOX-5 | | Authors: | Cary, P.D, Read, C.M, Davis, B, Driscoll, P.C, Crane-Robinson, C. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5.
Protein Sci., 10, 2001
|
|
1HSM
 
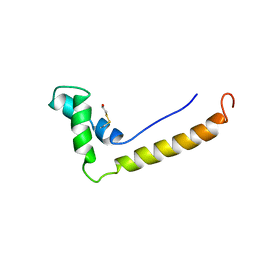 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
7X5P
 
 | | Truncated VhChiP (1-19aa) in complex with doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, ... | | Authors: | Sanram, S, Robinson, C.R, Aunkham, A, Suginta, W. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of truncated VhChiP
To Be Published
|
|
6RLL
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ44064146 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-5-(1,8-diazaspiro[4.5]decan-1-ylmethyl)oxolane-3,4-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A chemical probe for the methyl transferase PRMT5 with a novel
binding mode
To Be Published
|
|
6RLQ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882 | | Descriptor: | (1~{S},2~{R},3~{S},5~{R})-3-[2-(2-azanyl-3-bromanyl-quinolin-7-yl)ethyl]-5-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)cyclop entane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882
To Be Published
|
|
7BO7
 
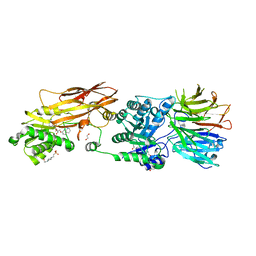 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJB44355437 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[(4~{a}~{S},7~{a}~{S})-4-azanyl-1,4,4~{a},7~{a}-tetrahydropyrrolo[2,3-d]pyrimidin-7-yl]-5-(quinolin-7-yloxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Brown, D, Robinson, C, Carr, K.H, Pande, V. | | Deposit date: | 2021-01-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJB44355437
To Be Published
|
|
1NHN
 
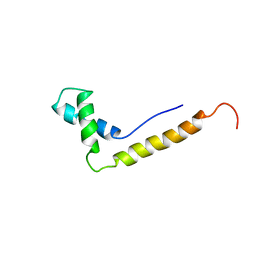 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NHM
 
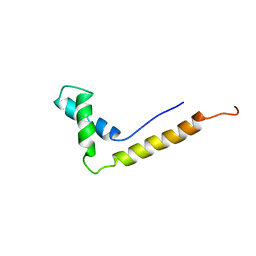 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
8OX0
 
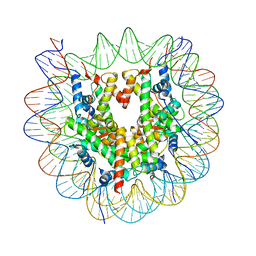 | | Structure of apo telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7JZ3
 
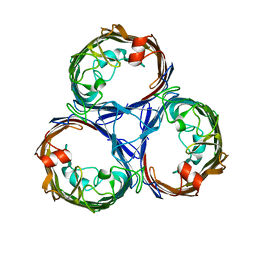 | | Osmoporin OmpC from E.coli K12 | | Descriptor: | Outer membrane protein C | | Authors: | Lyu, M, Su, C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7JZ2
 
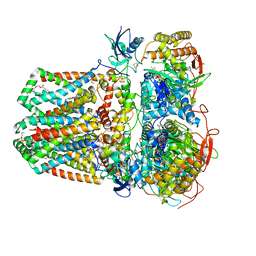 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
8OX1
 
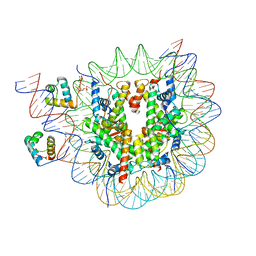 | | Structure of TRF1core in complex with telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
6O7T
 
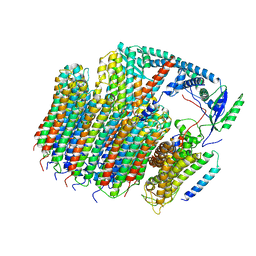 | | Saccharomyces cerevisiae V-ATPase Vph1-VO | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7V
 
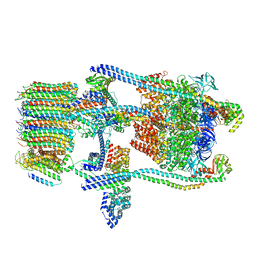 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8ARJ
 
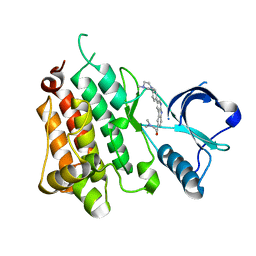 | | Anaplastic Lymphoma Kinase with a novel carboline inhibitor | | Descriptor: | 3-(dimethylamino)-1-[3-[4-(4-methylpiperazin-1-yl)phenyl]-9~{H}-pyrido[2,3-b]indol-6-yl]prop-2-en-1-one, ALK tyrosine kinase receptor | | Authors: | Mologni, L, Scapozza, L, Gambacorti-Passerini, C, Tardy, S, Goekjian, P, Robinson, C, Brown, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Discovery of Novel alpha-Carboline Inhibitors of the Anaplastic Lymphoma Kinase.
Acs Omega, 7, 2022
|
|
6O7W
 
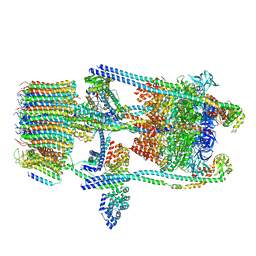 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 2 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
