4CHV
 
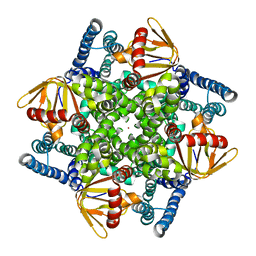 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-Induced Structural Changes in the Cyclic Nucleotide-Modulated Potassium Channel Mlok1
Nat.Commun., 5, 2014
|
|
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TG2
 
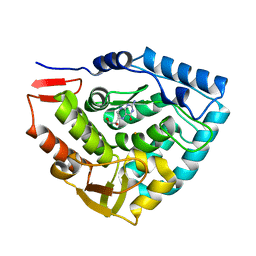 | | Crystal structure of phenylalanine hydroxylase A313T mutant with 7,8-dihydrobiopterin bound | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4NY7
 
 | |
4PAC
 
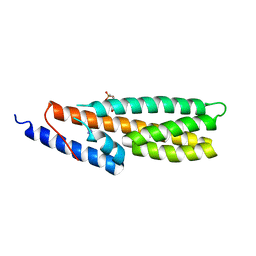 | | Crystal Structure of Histidine-containing Phosphotransfer Protein AHP2 from Arabidopsis thaliana | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Histidine-containing phosphotransfer protein 2, IMIDAZOLE | | Authors: | Degtjarik, O, Dopitova, R, Puehringer, S, Weiss, M.S, Janda, L, Hejatko, J, Kuta-Smatanova, I. | | Deposit date: | 2014-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of AHP2 from Arabidopsis thaliana
To Be Published
|
|
6TDQ
 
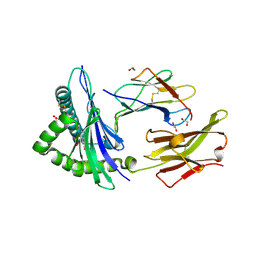 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket and one GM dipeptide in the F pocket. | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDS
 
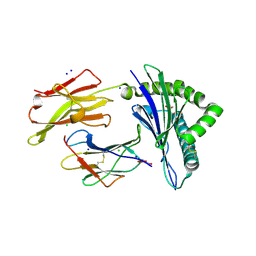 | | Crystal structure of the disulfide engineered HLA-A0201 molecule without peptide bound after NaCl wash | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDP
 
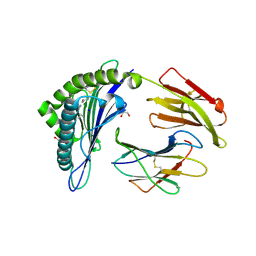 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GL dipeptide in the A pocket. | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDR
 
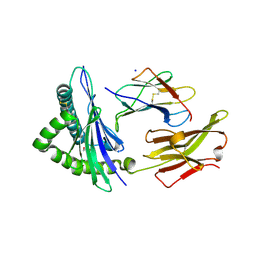 | | Crystal structure of the disulfide engineered HLA-A0201 molecule devoid of peptide (annealed) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
6TDO
 
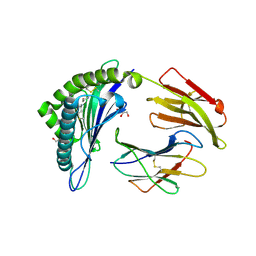 | | Crystal structure of the disulfide engineered HLA-A0201 molecule in complex with one GM dipeptide in the A pocket. | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Anjanappa, R, Garcia Alai, M, Springer, S, Meijers, R. | | Deposit date: | 2019-11-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection.
Nat Commun, 11, 2020
|
|
4EWI
 
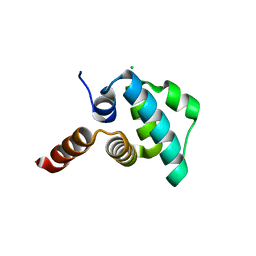 | | Crystal structure of the NLRP4 Pyrin domain | | Descriptor: | CHLORIDE ION, NACHT, LRR and PYD domains-containing protein 4, ... | | Authors: | Eibl, C, Hessenberger, M, Puehringer, S, Page, R, Diederichs, K, Peti, W. | | Deposit date: | 2012-04-27 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and Functional Analysis of the NLRP4 Pyrin Domain.
Biochemistry, 51, 2012
|
|
4HS3
 
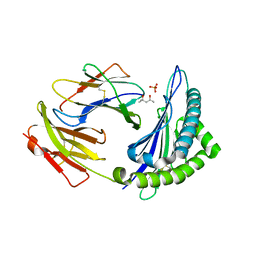 | | Crystal structure of H-2Kb with a disulfide stabilized F pocket in complex with the LCMV derived peptide GP34 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, Envelope glycoprotein, ... | | Authors: | Uchtenhagen, H, Boulanger, B, Hein, Z, Abualrous, E.T, Zacharias, M, Werner, J, Elliott, T, Springer, S, Achour, A. | | Deposit date: | 2012-10-29 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide-independent stabilization of MHC class I molecules breaches cellular quality control.
J.Cell.Sci., 127, 2014
|
|
4H1U
 
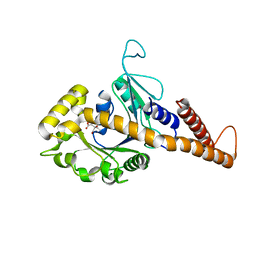 | | Nucleotide-free human dynamin-1-like protein GTPase-GED fusion | | Descriptor: | CITRATE ANION, Dynamin-1-like protein | | Authors: | Wenger, J, Klinglmayr, E, Puehringer, S, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
2I68
 
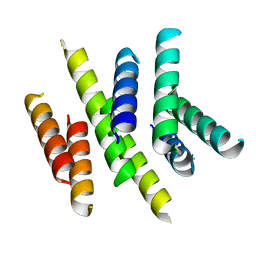 | | Cryo-EM based theoretical model structure of transmembrane domain of the multidrug-resistance antiporter from E. coli EmrE | | Descriptor: | Protein emrE | | Authors: | Fleishman, S.J, Harrington, S.E, Enosh, A, Halperin, D, Tate, C.G, Ben-Tal, N. | | Deposit date: | 2006-08-28 | | Release date: | 2006-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (7.5 Å) | | Cite: | Quasi-symmetry in the Cryo-EM Structure of EmrE Provides the Key to Modeling its Transmembrane Domain
J.Mol.Biol., 364, 2006
|
|
1KD5
 
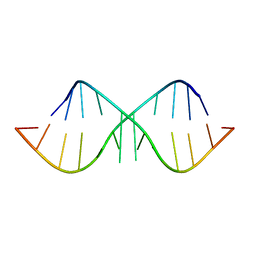 | | The Crystal Structure of r(GGUCACAGCCC)2 metal free form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3' | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KD3
 
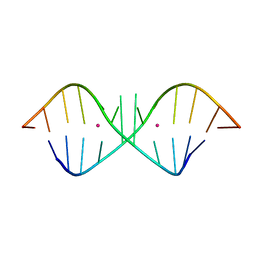 | | The Crystal Structure of r(GGUCACAGCCC)2, Thallium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', THALLIUM (I) ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KD4
 
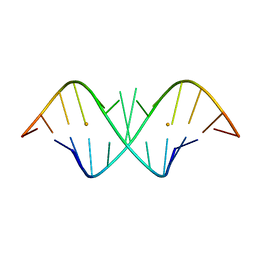 | | The Crystal Structure of r(GGUCACAGCCC)2, Barium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', BARIUM ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
