3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | 著者 | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | 登録日 | 2011-04-20 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
5OEM
 
 | |
3RLI
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | 著者 | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | 登録日 | 2011-04-19 | | 公開日 | 2012-05-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.854 Å) | | 主引用文献 | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
5OEN
 
 | | Crystal Structure of STAT2 in complex with IRF9 | | 分子名称: | Interferon regulatory factor 9, Signal transducer and activator of transcription | | 著者 | Rengachari, S, Panne, D. | | 登録日 | 2017-07-09 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.919 Å) | | 主引用文献 | Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | 分子名称: | Non-template strand, TATA-box-binding protein, Template strand, ... | | 著者 | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | 登録日 | 2022-05-20 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | 登録日 | 2022-05-20 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | 登録日 | 2022-05-20 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZWD
 
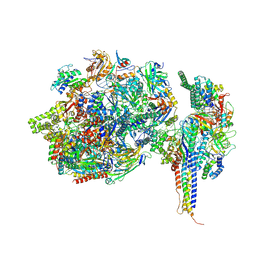 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | 登録日 | 2022-05-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8RAN
 
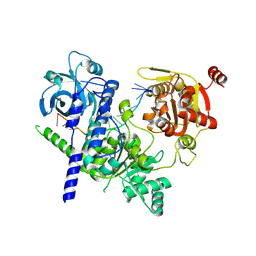 | |
8RAO
 
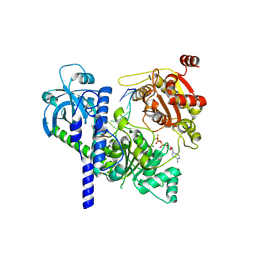 | |
4KE8
 
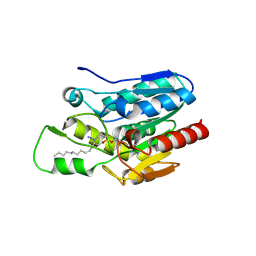 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | 分子名称: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | 著者 | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | 登録日 | 2013-04-25 | | 公開日 | 2013-09-18 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | 分子名称: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | 著者 | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | 登録日 | 2013-04-25 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE9
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-stearyol glycerol analogue | | 分子名称: | Thermostable monoacylglycerol lipase, hexadecyl hydrogen (R)-(3-azidopropyl)phosphonate | | 著者 | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | 登録日 | 2013-04-25 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
8RAM
 
 | |
8RAP
 
 | |
7ZWC
 
 | | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | | 分子名称: | Non-template strand, TATA-box-binding protein, Template strand, ... | | 著者 | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | 登録日 | 2022-05-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4KEA
 
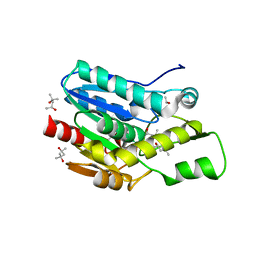 | | Crystal structure of D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in space group P212121 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | 著者 | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | 登録日 | 2013-04-25 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE7
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-myristoyl glycerol analogue | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase, dodecyl hydrogen (S)-(3-azidopropyl)phosphonate | | 著者 | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | 登録日 | 2013-04-25 | | 公開日 | 2013-09-18 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
7NVR
 
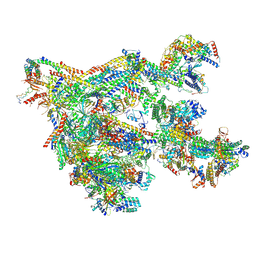 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | 登録日 | 2021-03-15 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
4ZWN
 
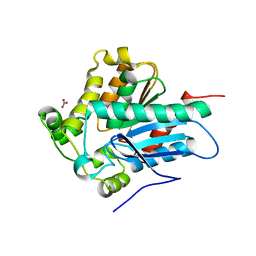 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | 分子名称: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | 著者 | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | 登録日 | 2015-05-19 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.491 Å) | | 主引用文献 | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
4ZXF
 
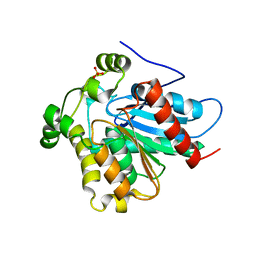 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | 分子名称: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | 著者 | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | 登録日 | 2015-05-20 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
6RSR
 
 | | TBK1 in complex with compound 2 | | 分子名称: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | 著者 | Panne, D, Hillig, R.C, Rengachari, S. | | 登録日 | 2019-05-22 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RST
 
 | | TBK1 in complex with inhibitor compound 24 | | 分子名称: | 1-[4-[(1~{R})-1-[2-[[5-[1-(cyclopropylmethyl)pyrazol-4-yl]-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]-3,3,3-tris(fluoranyl)propan-1-one, Serine/threonine-protein kinase TBK1 | | 著者 | Panne, D, Hillig, R.C, Rengachari, S. | | 登録日 | 2019-05-22 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RSU
 
 | | TBK1 in complex with Inhibitor compound 35 | | 分子名称: | 3,3,3-tris(fluoranyl)-1-[4-[(1~{R})-1-[2-[[(2~{S})-5-(5-propan-2-yloxypyrimidin-4-yl)-2,3-dihydro-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]propan-1-one, Serine/threonine-protein kinase TBK1 | | 著者 | Panne, D, Hillig, R.C, Rengachari, S. | | 登録日 | 2019-05-22 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
