7C1S
 
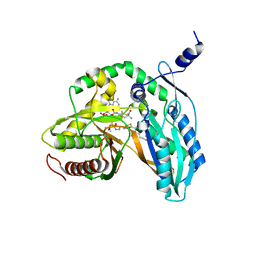 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA and Leu-SNAC | | Descriptor: | Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A, S-(2-acetamidoethyl) (2S)-2-azanyl-4-methyl-pentanethioate | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1K
 
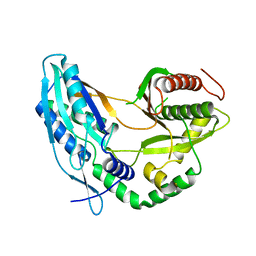 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1L
 
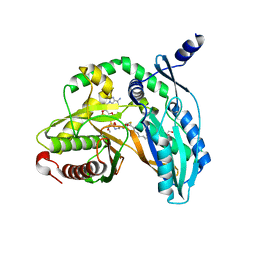 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1U
 
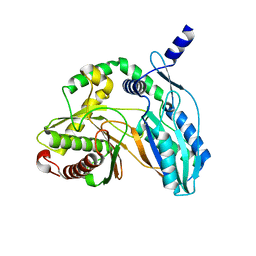 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140V/R148A in a "product-released" conformation | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
4TQK
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQJ
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQM
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4RR4
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367 | | Descriptor: | 2-chloro-N-[3-(4-{[(2Z)-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enoyl]amino}phenoxy)phenyl]-4-methyl-1,3-thiazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-11-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367
TO BE PUBLISHED
|
|
4RKA
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347 | | Descriptor: | 2-{[5-(naphthalen-1-ylmethyl)-4-oxo-4H-1lambda~4~,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347
TO BE PUBLISHED
|
|
4RSN
 
 | | Crystal structure of the E267V mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W.H, Zhang, A.L, Zhang, T, Ren, X.K, Yorke, J.A, Taylor, A, Wong, L.-L. | | Deposit date: | 2014-11-09 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Expanding the drug metabolism function of P450BM3
To be Published
|
|
4RK8
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356 | | Descriptor: | 5-fluoro-2-{[(5Z)-5-(naphthalen-1-ylmethylidene)-4-oxo-4,5-dihydro-1,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356
TO BE PUBLISHED
|
|
4RLI
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048 | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048
TO BE PUBLISHED
|
|
7V7W
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7V7L
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7JTL
 
 | | Structure of SARS-CoV-2 ORF8 accessory protein | | Descriptor: | ORF8 protein, SODIUM ION | | Authors: | Flower, T.G, Buffalo, C.Z, Hooy, R.M, Allaire, M, Ren, X, Hurley, J.H. | | Deposit date: | 2020-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XSV
 
 | | Crystal Structures of PIM1 in Complex with Macrocyclic Compound H3 | | Descriptor: | 8-Methyl-2,5,20-trioxa-8,13,17-triazatetracyclo[11.10.2.014,19.021,25]pentacosa-1(24),14(19),15,17,21(25),22-hexaene, Serine/threonine-protein kinase pim-1 | | Authors: | Shen, C, Xie, Y, Ren, X, Zhou, Y, Niu, H. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Design, synthesis, and bioactivity evaluation of macrocyclic benzo[b]pyrido[4,3-e][1,4]oxazine derivatives as novel Pim-1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 72, 2022
|
|
6CM9
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef closed trimer monomeric subunit | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-03-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
6DFF
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef monomer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
5HM9
 
 | | Crystal structure of MamO protease domain from Magnetospirillum magneticum (apo form) | | Descriptor: | MamO protease domain, poly(UNK) | | Authors: | Hershey, D.M, Ren, X, Hurley, J.H, Komeili, A. | | Deposit date: | 2016-01-15 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Plos Biol., 14, 2016
|
|
5HMA
 
 | | Crystal structure of MamO protease domain from Magnetospirillum magneticum (Ni bound form) | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Trypsin-like serine protease, ... | | Authors: | Hershey, D.M, Ren, X, Hurley, J.H, Komeili, A. | | Deposit date: | 2016-01-15 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Plos Biol., 14, 2016
|
|
4LS1
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A312 | | Descriptor: | 2-[(E)-{2-[4-(2-chlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}methyl]benzoic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A312
To be Published
|
|
4LS2
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A313 | | Descriptor: | 2-[(E)-{2-[4-(3-methoxyphenyl)-1,3-thiazol-2-yl]hydrazinylidene}methyl]benzoic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A313
To be Published
|
|
4JGD
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016 | | Descriptor: | 1-[4-methyl-2-(naphthalen-2-ylamino)-1,3-thiazol-5-yl]ethanone, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016
TO BE PUBLISHED
|
|
4JTT
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 066 | | Descriptor: | 2,4-dimethyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
4NEE
 
 | | crystal structure of AP-2 alpha/simga2 complex bound to HIV-1 Nef | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit sigma, Protein Nef | | Authors: | Hurley, J.H, Bonifacino, J.S, Ren, X, Park, S.Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8841 Å) | | Cite: | How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4.
Elife, 3, 2014
|
|
