1UXW
 
 | | CRYSTAL STRUCTURE OF HLA-B*2709 COMPLEXED WITH THE LATENT MEMBRANE PROTEIN 2 PEPTIDE (LMP2) OF EPSTEIN-BARR VIRUS | | Descriptor: | BETA-2-MICROGLOBULIN, GENE TERMINAL PROTEIN (MEMBRANE PROTEIN LMP-2A/LMP-2B), GLYCEROL, ... | | Authors: | Hulsmeyer, M, Kozerski, C, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-03-01 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Allele-Dependent Similarity between Viral and Self-Peptide Presentation by Hla-B27 Subtypes
J.Biol.Chem., 280, 2005
|
|
1UXS
 
 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE LATENT MEMBRANE PROTEIN 2 PEPTIDE (LMP2)OF EPSTEIN-BARR VIRUS | | Descriptor: | BETA-2-MICROGLOBULIN, GENE TERMINAL PROTEIN (MEMBRANE PROTEIN LMP-2A/LMP-2B), GLYCEROL, ... | | Authors: | Hulsmeyer, M, Kozerski, C, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-03-01 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Allele-Dependent Similarity between Viral and Self-Peptide Presentation by Hla-B27 Subtypes
J.Biol.Chem., 280, 2005
|
|
1SU5
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1W0V
 
 | | Crystal Structure Of HLA-B*2705 Complexed With the self-Peptide TIS from EGF-response factor 1 | | Descriptor: | BETA-2-MICROGLOBULIN, BUTYRATE RESPONSE FACTOR 2, GLYCEROL, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-06-14 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Thermodynamic and Structural Equivalence of Two Hla-B27 Subtypes Complexed with a Self-Peptide
J.Mol.Biol., 346, 2005
|
|
1M3Z
 
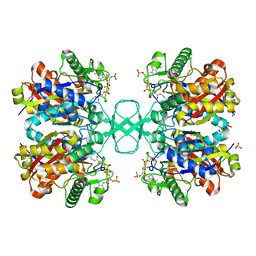 | | Biosynthetic thiolase, C89A mutant, complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1TJC
 
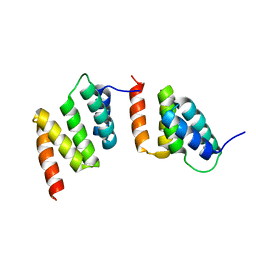 | | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | | Descriptor: | Prolyl 4-hydroxylase alpha-1 subunit | | Authors: | Pekkala, M, Hieta, R, Bergmann, U, Kivirikko, K.I, Wierenga, R.K, Myllyharju, J. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Peptide-Substrate-binding Domain of Collagen Prolyl 4-Hydroxylases Is a Tetratricopeptide Repeat Domain with Functional Aromatic Residues.
J.Biol.Chem., 279, 2004
|
|
1M1T
 
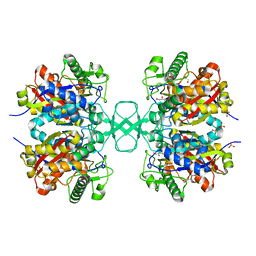 | | Biosynthetic thiolase, Q64A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M4S
 
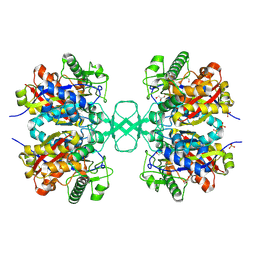 | | Biosynthetic thiolase, Cys89 acetylated, unliganded form | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M1O
 
 | | Crystal structure of biosynthetic thiolase, C89A mutant, complexed with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M4T
 
 | | Biosynthetic thiolase, Cys89 butyrylated | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
2GD0
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (S)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
1SQ7
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1M3K
 
 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
2J24
 
 | | The functional role of the conserved active site proline of triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Casteleijn, M.G, Alahuhta, M, Groebel, K, El-Sayed, I, Augustyns, K, Lambeir, A.M, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2006-08-16 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional Role of the Conserved Active Site Proline of Triosephosphate Isomerase.
Biochemistry, 45, 2006
|
|
1WL4
 
 | | Human cytosolic acetoacetyl-CoA thiolase complexed with CoA | | Descriptor: | COENZYME A, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, P, Fukao, T, Kondo, N, Wierenga, R.K. | | Deposit date: | 2004-06-20 | | Release date: | 2005-03-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High Resolution Crystal Structures of Human Cytosolic Thiolase (CT): A Comparison of the Active Sites of Human CT, Bacterial Thiolase, and Bacterial KAS I
J.Mol.Biol., 347, 2005
|
|
1SSD
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1WL5
 
 | | Human cytosolic acetoacetyl-CoA thiolase | | Descriptor: | GLYCEROL, SULFATE ION, acetyl-Coenzyme A acetyltransferase 2 | | Authors: | Kursula, P, Fukao, T, Kondo, N, Wierenga, R.K. | | Deposit date: | 2004-06-20 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High Resolution Crystal Structures of Human Cytosolic Thiolase (CT): A Comparison of the Active Sites of Human CT, Bacterial Thiolase, and Bacterial KAS I
J.Mol.Biol., 347, 2005
|
|
2RNF
 
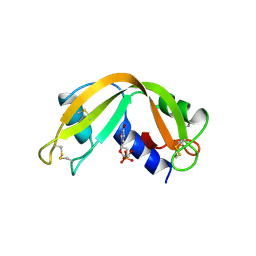 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
3IAO
 
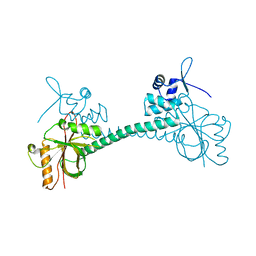 | |
2JF4
 
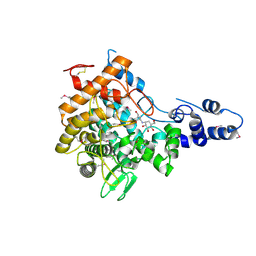 | | Family 37 trehalase from Escherichia coli in complex with validoxylamine | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, PERIPLASMIC TREHALASE | | Authors: | Gibson, R.P, Gloster, T.M, Roberts, S, Warren, R.A.J, Storch De Gracia, I, Garcia, A, Chiara, J.L, Davies, G.J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for Trehalase Inhibition Revealed by the Structure of Trehalase in Complex with Potent Inhibitors.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
1OGT
 
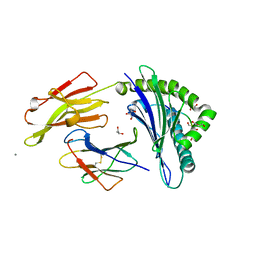 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE VASOACTIVE INTESTINAL PEPTIDE TYPE 1 RECEPTOR (VIPR) PEPTIDE (RESIDUES 400-408) | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2003-05-13 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dual, HLA-B27 subtype-dependent conformation of a self-peptide.
J. Exp. Med., 199, 2004
|
|
1ODZ
 
 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
2VCY
 
 | | Crystal Structure of 2-Enoyl Thioester Reductase of Human FAS II | | Descriptor: | SULFATE ION, TRANS-2-ENOYL-COA REDUCTASE | | Authors: | Haapalainen, A.M, Pudas, R, Smart, O.S, Wierenga, R.K. | | Deposit date: | 2007-09-28 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Enzymological Studies of 2-Enoyl Thioester Reductase of the Human Mitochondrial Fas II Pathway: New Insights Into its Substrate Recognition Properties.
J.Mol.Biol., 379, 2008
|
|
1O8P
 
 | | Unbound structure of CsCBM6-3 from Clostridium stercorarium | | Descriptor: | CALCIUM ION, PUTATUVE ENDO-XYLANASE | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains.
J. Mol. Biol., 327, 2003
|
|
1SPQ
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
