1AHA
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ADENINE, ALPHA-MOMORCHARIN | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1AHB
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ALPHA-MOMORCHARIN, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1JLF
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLE
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | HIV-1 RT, A-CHAIN, B-CHAIN | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLA
 
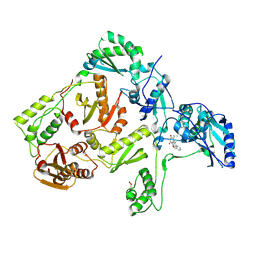 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JKH
 
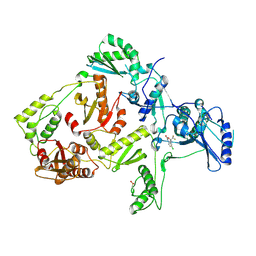 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLG
 
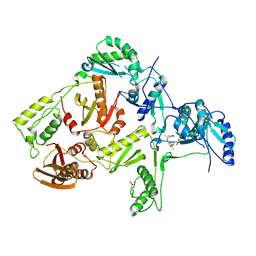 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLB
 
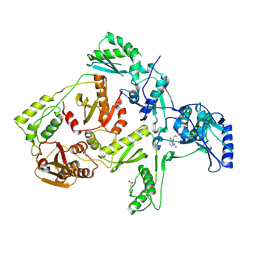 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLQ
 
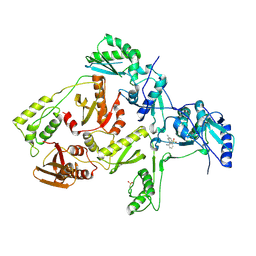 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
1JLC
 
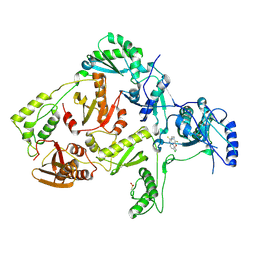 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
4G2W
 
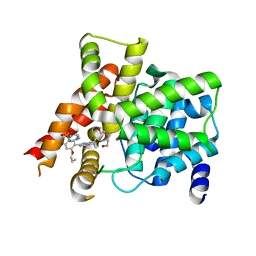 | | Crystal structure of PDE5A in complex with its inhibitor | | Descriptor: | 5,6-diethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-07-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
4G2Y
 
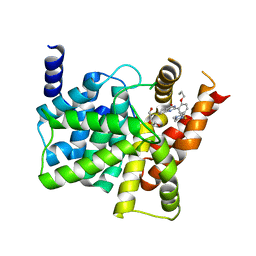 | | Crystal structure of PDE5A complexed with its inhibitor | | Descriptor: | 2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-07-13 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
4YU8
 
 | | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Ren, J, Nettleship, J.E, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2015-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1
To Be Published
|
|
1LW2
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWC
 
 | | CRYSTAL STRUCTURE OF M184V MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWE
 
 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
2A0J
 
 | | Crystal Structure of Nitrogen Regulatory Protein IIA-Ntr from Neisseria meningitidis | | Descriptor: | PTS system, nitrogen regulatory IIA protein | | Authors: | Ren, J, Sainsbury, S, Berrow, N.S, Alderton, D, Nettleship, J.E, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2005-06-16 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis
Bmc Struct.Biol., 5, 2005
|
|
4JGZ
 
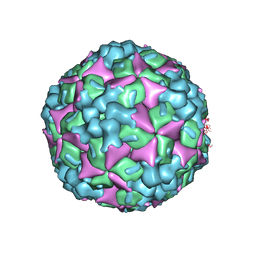 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4JGY
 
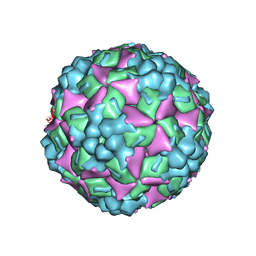 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
1FKO
 
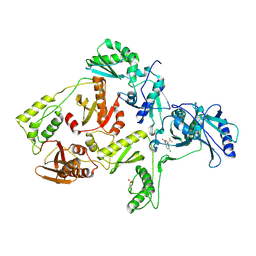 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1FK9
 
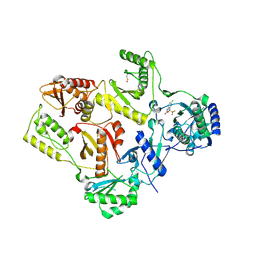 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-09 | | Release date: | 2000-11-03 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
STRUCTURE FOLD.DES., 8, 2000
|
|
1FKP
 
 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1EP4
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with S-1153 | | Descriptor: | 5-(3,5-DICHLOROPHENYL)THIO-4-ISOPROPYL-1-(PYRIDIN-4-YL-METHYL)-1H-IMIDAZOL-2-YL-METHYL CARBAMATE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Nichols, C, Bird, L.E, Fujiwara, T, Suginoto, H, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-03-27 | | Release date: | 2000-09-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding.
J.Biol.Chem., 275, 2000
|
|
