5KCB
 
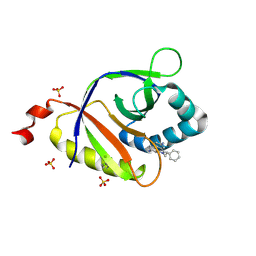 | | The structure of SAV2435 bound to ethidium bromide | | Descriptor: | ETHIDIUM, SA2223 protein, SULFATE ION | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
4O9H
 
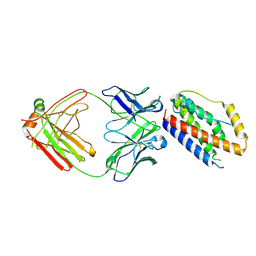 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | Descriptor: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | Authors: | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | Deposit date: | 2014-01-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
3QI8
 
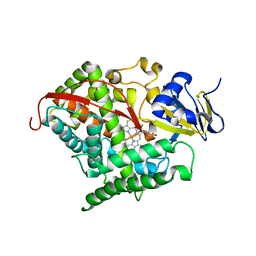 | | Evolved variant of cytochrome P450 (BM3, CYP102A1) | | Descriptor: | Evolved Cytochrome P450 variant (22A3), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rentmeister, A, Brown, T.R, Snow, C.D, Carbone, M.N, Arnold, F.H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineered Bacterial Mimics of Human Drug Metabolizing Enzyme CYP2C9
Chemcatchem, 2011
|
|
3SDJ
 
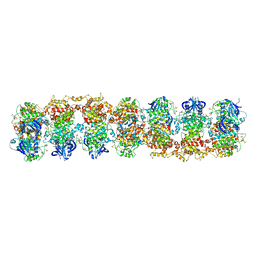 | | Structure of RNase-inactive point mutant of oligomeric kinase/RNase Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and functional basis for RNA cleavage by Ire1.
Bmc Biol., 9, 2011
|
|
3SDM
 
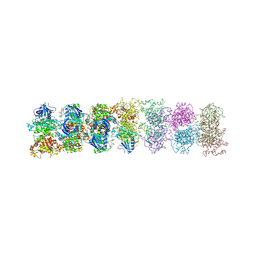 | | Structure of oligomeric kinase/RNase Ire1 in complex with an oligonucleotide | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1.
Bmc Biol., 9, 2011
|
|
3DPA
 
 | |
1QL4
 
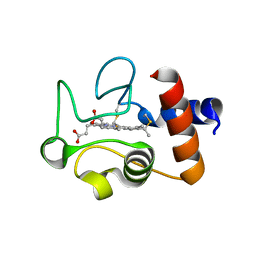 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
2ESG
 
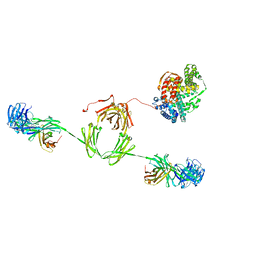 | | Solution structure of the complex between immunoglobulin IgA1 and human serum albumin | | Descriptor: | Immunoglobulin A1 heavy chain, Immunoglobulin A1 light chain, Serum albumin | | Authors: | Almogren, A, Furtado, P.B, Sun, Z, Perkins, S.J, Kerr, M.A. | | Deposit date: | 2005-10-26 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Purification, Properties and Extended Solution Structure of the Complex Formed between Human Immunoglobulin A1 and Human Serum Albumin by Scattering and Ultracentrifugation.
J.Mol.Biol., 356, 2006
|
|
1QL3
 
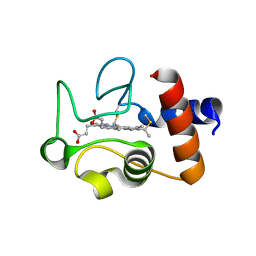 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the reduced state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QLE
 
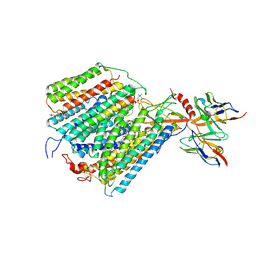 | |
6LAX
 
 | | the mutant SAM-VI riboswitch (U6C) bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Sun, A, Ren, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
5Y87
 
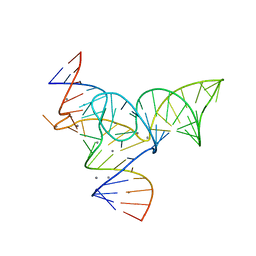 | |
7W27
 
 | | Crystal structure of BEND3-BEN4-DNA complex | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*CP*GP*TP*GP*GP*GP*TP*C)-3') | | Authors: | Zheng, L, Ren, A. | | Deposit date: | 2021-11-22 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins.
Genes Dev., 36, 2022
|
|
5Y85
 
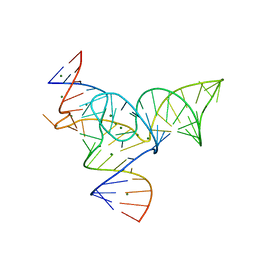 | |
1U4R
 
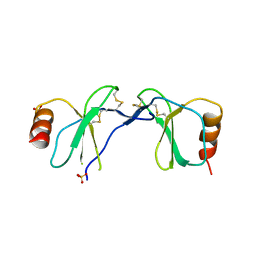 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4L
 
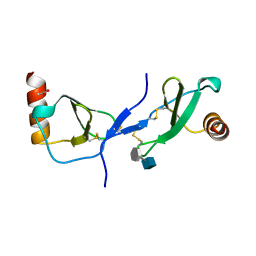 | | human RANTES complexed to heparin-derived disaccharide I-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4M
 
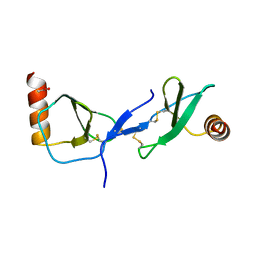 | | human RANTES complexed to heparin-derived disaccharide III-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4P
 
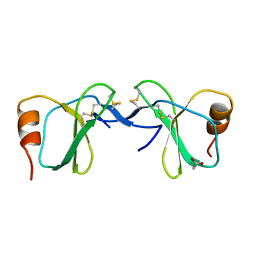 | | Crystal Structure of human RANTES mutant K45E | | Descriptor: | ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
8QPH
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra 14 crystals | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | VMXm - sub-micron microfocus beamline for macromolecular crystallography at Diamond Light Source
To Be Published
|
|
8QQC
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra single crystal | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | VMXm - sub-micron microfocus beamline for macromolecular crystallography at Diamond Light Source
To Be Published
|
|
4D4H
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4I
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, ARGININE, GLYCEROL, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4G
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | APNAA1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D57
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNA A1, ARGININE, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D56
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNAA1, GLYCEROL, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
