5MRH
 
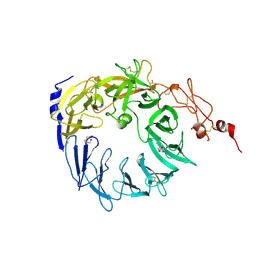 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-methylbutyl)-4~{H}-1,2,3-triazol-5-one, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4O4R
 
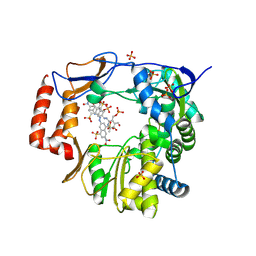 | | Murine Norovirus RdRp in complex with PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | Authors: | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
4NRU
 
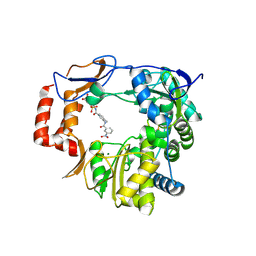 | | Murine Norovirus RNA-dependent-RNA-polymerase in complex with Compound 6, a suramin derivative | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, MAGNESIUM ION, RNA dependent RNA polymerase | | Authors: | Milani, M, Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
6V7B
 
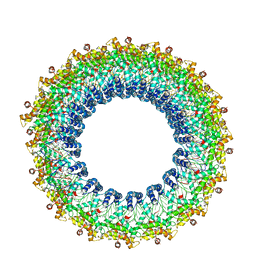 | | Cryo-EM reconstruction of Pyrobaculum filamentous virus 2 (PFV2) | | Descriptor: | A-DNA, Structural protein VP1, Structural protein VP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2019-12-08 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Virus Evol, 6, 2020
|
|
1ZSN
 
 | | Synthesis, Biological Activity, and X-Ray Crystal Structural Analysis of Diaryl Ether Inhibitors of Malarial Enoyl ACP Reductase. Part 1:4'-Substituted Triclosan Derivatives | | Descriptor: | 5-CHLORO-2-(2-CHLORO-4-NITROPHENOXY)PHENOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, enoyl-acyl carrier reductase | | Authors: | Freundlich, J.S, Anderson, J.W, Sarantakis, D, Shieh, H.M, Yu, M, Lucumi, E, Kuo, M, Schiehser, G.A, Jacobus, D.P, Jacobs Jr, W.R, Fidock, D.A, Sacchettini, J.C. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Synthesis, biological activity, and X-ray crystal structural analysis of diaryl ether inhibitors of malarial enoyl acyl carrier protein reductase. Part 1: 4'-Substituted triclosan derivatives.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2QVC
 
 | | Crystal structure of a periplasmic sugar ABC transporter from Thermotoga maritima | | Descriptor: | Sugar ABC transporter, periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
2QXY
 
 | |
1ZXL
 
 | | Synthesis, Biological Activity, and X-Ray Crystal Structural Analysis of Diaryl Ether Inhibitors of Malarial Enoyl ACP Reductase. Part 1:4'-Substituted Triclosan Derivatives | | Descriptor: | N-[3-CHLORO-4-(4-CHLORO-2-HYDROXYPHENOXY)PHENYL]MORPHOLINE-4-CARBOXAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, enoyl-acyl carrier reductase | | Authors: | Freundlich, J.S, Anderson, J.W, Sarantakis, D, Shieh, H.M, Yu, M, Lucumi, E, Kuo, M, Schiehser, G.A, Jacobus, D.P, Jacobs Jr, W.R, Fidock, D.A, Sacchettini, J.C. | | Deposit date: | 2005-06-08 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, biological activity, and X-ray crystal structural analysis of diaryl ether inhibitors of malarial enoyl acyl carrier protein reductase. Part 1: 4'-Substituted triclosan derivatives.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5W3K
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex NADPH, Mg2+ and CPD | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Kandale, A, Schembri, M, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
4NRT
 
 | | Human Norovirus polymerase bound to Compound 6 (suramin derivative) | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, hNV-RdRp | | Authors: | Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Milani, M, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
5OC0
 
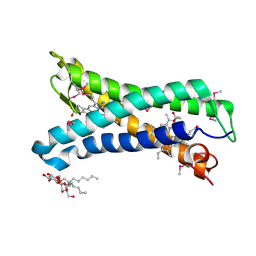 | | Structure of E. coli superoxide oxidase | | Descriptor: | Cytochrome b561, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lundgren, C.A.K, Sjostrand, D, Biner, O, Bennett, M, von Ballmoos, C, Hogbom, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Scavenging of superoxide by a membrane-bound superoxide oxidase.
Nat. Chem. Biol., 14, 2018
|
|
7K6E
 
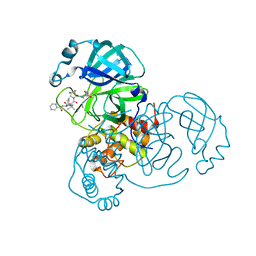 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
1PV1
 
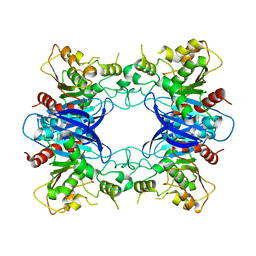 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | Descriptor: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | Authors: | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-26 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
7JYC
 
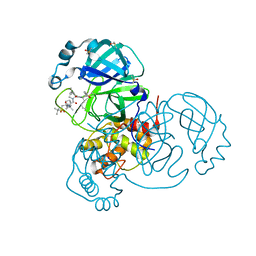 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
5NF8
 
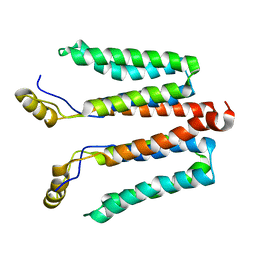 | | Solution structure of detergent-solubilized Rcf1, a yeast mitochondrial inner membrane protein involved in respiratory Complex III/IV supercomplex formation | | Descriptor: | Respiratory supercomplex factor 1, mitochondrial | | Authors: | Zhou, S, Pettersson, P, Sjoholm, J, Sjostrand, D, Hogbom, M, Brzezinski, P, Maler, L, Adelroth, P. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast Rcf1, a protein involved in respiratory supercomplex formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MHL
 
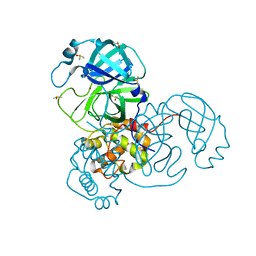 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
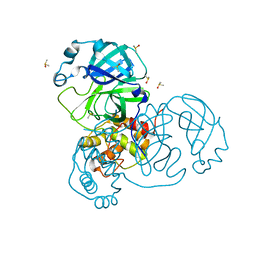 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7JNY
 
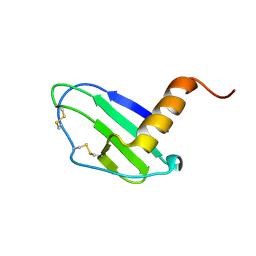 | | Crystal structure of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Rajasekaran, D, Murphy, J.W, Pantouris, G, Lolis, E.J. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1I1E
 
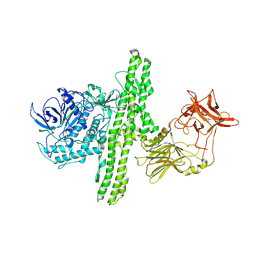 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM BOTULINUM NEUROTOXIN B COMPLEXED WITH DOXORUBICIN | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, DOXORUBICIN, SULFATE ION, ... | | Authors: | Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2001-02-01 | | Release date: | 2001-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for doxorubicin binding to the receptor-binding site in Clostridium botulinum neurotoxin B.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5W7G
 
 | | An envelope of a filamentous hyperthermophilic virus carries lipids in a horseshoe conformation | | Descriptor: | DNA (253-MER), ORF132, ORF140 | | Authors: | Kasson, P, DiMaio, F, Yu, X, Lucas-Staat, S, Krupovic, M, Schouten, S, Prangishvili, D, Egelman, E. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Model for a novel membrane envelope in a filamentous hyperthermophilic virus.
Elife, 6, 2017
|
|
2WB6
 
 | | Crystal structure of AFV1-102, a protein from the Acidianus Filamentous Virus 1 | | Descriptor: | AFV1-102, CHLORIDE ION | | Authors: | Keller, J, Leulliot, N, Collinet, B, Campanacci, V, Cambillau, C, Pranghisvilli, D, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Afv1-102, a Protein from the Acidianus Filamentous Virus 1.
Protein Sci., 18, 2009
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
