6R1T
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | 分子名称: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.02 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.61 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6NTA
 
 | | Modified ASL proline bound to Thermus thermophilus 70S (cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Maehigashi, T, Subaramanian, S, Hong, S, Dunham, C.M. | | 登録日 | 2019-01-28 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6NWY
 
 | | Modified tRNA(Pro) bound to Thermus thermophilus 70S (near-cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | 登録日 | 2019-02-07 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6NSH
 
 | | Modified ASL proline bound to Thermus thermophilus 70S (near-cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Maehigashi, T, Subaramanian, S, Hong, S, Dunham, C.M. | | 登録日 | 2019-01-24 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.397 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6O3M
 
 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | 登録日 | 2019-02-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.97 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6OSI
 
 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (near cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | 登録日 | 2019-05-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (4.14 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6NUO
 
 | | Modified tRNA(Pro) bound to Thermus thermophilus 70S (cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | 登録日 | 2019-02-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | 分子名称: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | 分子名称: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-19 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | 分子名称: | Hepatitis C virus, ZINC ION | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-19 | | 公開日 | 2007-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6O0X
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | 分子名称: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | 著者 | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | 登録日 | 2019-02-17 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O0Z
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, non-target strand DNA, single guide RNA, ... | | 著者 | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | 登録日 | 2019-02-17 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O0Y
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | 分子名称: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | 著者 | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | 登録日 | 2019-02-17 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6X1Q
 
 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | 分子名称: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | 著者 | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | 登録日 | 2020-05-19 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (1.8 Å) | | 主引用文献 | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
6UAN
 
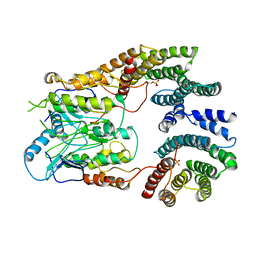 | | B-Raf:14-3-3 complex | | 分子名称: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | 著者 | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | 登録日 | 2019-09-11 | | 公開日 | 2019-09-25 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
6PWE
 
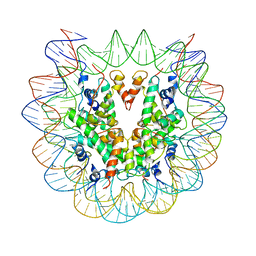 | |
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJI
 
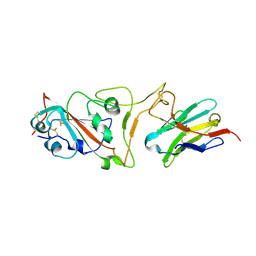 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJH
 
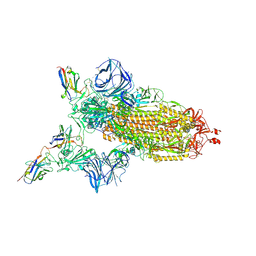 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
