2GSL
 
 | | X-Ray Crystal Structure of Protein FN1578 from Fusobacterium nucleatum. Northeast Structural Genomics Consortium Target NR1. | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Conover, K, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (FN1578) from Fusobacterium nucleatum, NESG Target NR1
To be Published
|
|
2GXF
 
 | | X-Ray Crystal Structure of Protein YybH from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR506. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein yybH | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Cunningham, K, Fang, Y, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-08 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein YybH from Bacillus subtilis, Northeast Structural Genomics Target SR506
TO BE PUBLISHED
|
|
2GWG
 
 | | Crystal Structure of 4-Oxalomesaconate Hydratase, LigJ, from Rhodopseudomonas palustris, Northeast Structural Genomics Target RpR66. | | Descriptor: | 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Cunningham, K, Ciao, M, Ma, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-04 | | Release date: | 2006-05-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 4-Oxalomesaconate Hydratase, LigJ, from Rhodopseudomonas palustris, Northeast Structural Genomics Target RpR66
To be Published
|
|
2G5V
 
 | |
2GF4
 
 | | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14 | | Descriptor: | ACETATE ION, CALCIUM ION, Protein Vng1086c | | Authors: | Benach, J, Zhou, W, Jayaraman, S, Forouhar, F.F, Janjua, H, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14
To be Published
|
|
2PK7
 
 | | Crystal structure of the Q4KFT4_PSEF5 protein from Pseudomonas fluorescens. NESG target PlR1 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Neely, H, Jayaraman, S, Chen, C.X, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Q4KFT4_PSEF5 protein from Pseudomonas fluorescens.
To be Published
|
|
2QGU
 
 | | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A. Northeast Structural Genomics Consortium target RsR89 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Probable signal peptide protein | | Authors: | Kuzin, A.P, Chen, Y, Jayaraman, S, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A.
To be Published
|
|
7LAD
 
 | | Clobetasol propionate bound to CYP3A5 | | Descriptor: | Clobetasol propionate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Buchman, C.D, Miller, D, Wang, J, Jayaraman, S, Chen, T. | | Deposit date: | 2021-01-06 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unraveling the Structural Basis of Selective Inhibition of Human Cytochrome P450 3A5.
J.Am.Chem.Soc., 143, 2021
|
|
2VDA
 
 | | Solution structure of the SecA-signal peptide complex | | Descriptor: | MALTOPORIN, TRANSLOCASE SUBUNIT SECA | | Authors: | Gelis, I, Bonvin, A.M.J.J, Keramisanou, D, Koukaki, M, Gouridis, G, Karamanou, S, Economou, A, Kalodimos, C.G. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Signal-Sequence Recognition by the Translocase Motor Seca as Determined by NMR
Cell(Cambridge,Mass.), 131, 2007
|
|
2QGM
 
 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
2QGG
 
 | | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248. Northeast Structural Genomics Consortium target AsR73. | | Descriptor: | 16S rRNA-processing protein rimM, POTASSIUM ION, UNKNOWN LIGAND | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248.
To be Published
|
|
2HD3
 
 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3JA7
 
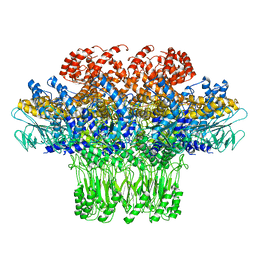 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
3JCZ
 
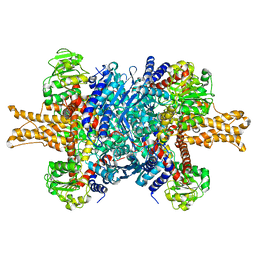 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
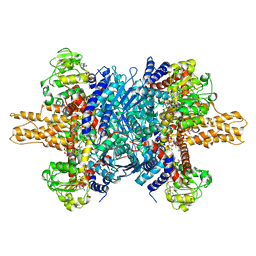 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3J7H
 
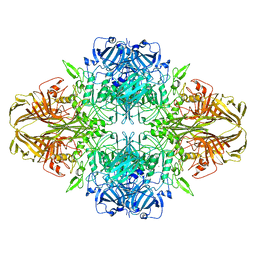 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JD2
 
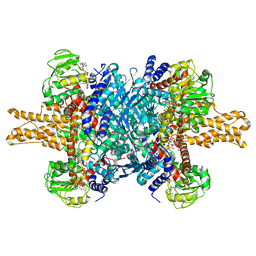 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
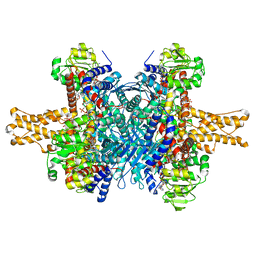 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JCH
 
 | |
3JCG
 
 | |
3JCF
 
 | |
3MXA
 
 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*C)-3'), DNA (5'-D(P*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
3MX9
 
 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
