2BCG
 
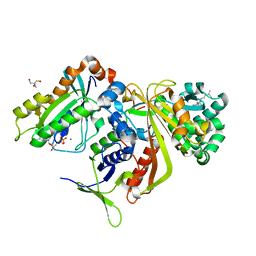 | | Structure of doubly prenylated Ypt1:GDI complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERAN-8-YL GERAN, GTP-binding protein YPT1, ... | | Authors: | Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2005-10-19 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling
Embo J., 25, 2006
|
|
3ZM4
 
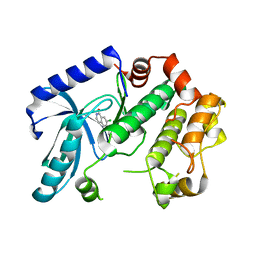 | | Crystal structure of MEK1 in complex with fragment 1 | | Descriptor: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ASZ
 
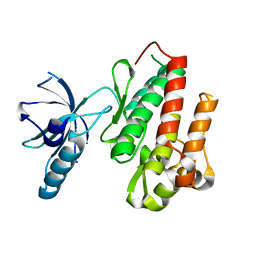 | | Crystal structure of apo TrkB kinase domain | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4CP6
 
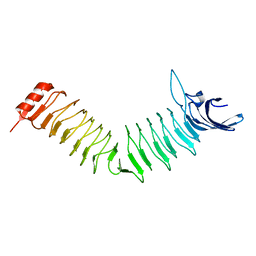 | | The Crystal structure of Pneumococcal vaccine antigen PcpA | | Descriptor: | CHOLINE BINDING PROTEIN PCPA | | Authors: | Vallee, F, Steier, V, Oloo, E, Chawla, D, Vonrhein, C, Steinmetz, A, Mathieu, M, Rak, A, Mikol, V, Oomen, R. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The Crystal Structure of Pneumoccocal Vaccine Antigen Pcpa
To be Published
|
|
1N88
 
 | | NMR structure of the ribosomal protein L23 from Thermus thermophilus. | | Descriptor: | Ribosomal protein L23 | | Authors: | Ohman, A, Rak, A, Dontsova, M, Garber, M.B, Hard, T. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the ribosomal protein L23 from Thermus thermophilus.
J.Biomol.NMR, 26, 2003
|
|
1QKH
 
 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S19 FROM THERMUS THERMOPHILUS | | Descriptor: | 30S RIBOSOMAL PROTEIN S19 | | Authors: | Helgstrand, M, Rak, A.V, Allard, P, Davydova, N, Garber, M.B, Hard, T. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal protein S19 from Thermus thermophilus.
J. Mol. Biol., 292, 1999
|
|
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLY
 
 | | Crystal structure of MEK1 in complex with fragment 8 | | Descriptor: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4AT5
 
 | | CRYSTAL STRUCTURE OF TRKB KINASE DOMAIN IN COMPLEX WITH GW2580 | | Descriptor: | 5-{3-methoxy-4-[(4-methoxybenzyl)oxy]benzyl}pyrimidine-2,4-diamine, BDNF/NT-3 GROWTH FACTORS RECEPTOR, GLYCEROL | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4AT3
 
 | | CRYSTAL STRUCTURE OF TRKB KINASE DOMAIN IN COMPLEX WITH CPD5N | | Descriptor: | (5Z)-5-(carbamoylimino)-3-[(5R)-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ylsulfanyl]-2,5-dihydroisothiazole-4-carboxamide, BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
7AZS
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-569 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,6R,E)-3-hydroxy-4-(methoxyimino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G, Rak, A. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
4AT4
 
 | | CRYSTAL STRUCTURE OF TRKB KINASE DOMAIN IN COMPLEX WITH EX429 | | Descriptor: | 1-[4-(4-aminothieno[2,3-d]pyrimidin-5-yl)phenyl]-3-[2-fluoro-5-(trifluoromethyl)phenyl]urea, BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
1ILY
 
 | | Solution Structure of Ribosomal Protein L18 of Thermus thermophilus | | Descriptor: | RIBOSOMAL PROTEIN L18 | | Authors: | Woestenenk, E.A, Gongadze, G.M, Shcherbakov, D.V, Rak, A.V, Garber, M.B, Hard, T, Berglund, H. | | Deposit date: | 2001-05-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold.
Biochem.J., 363, 2002
|
|
5FHX
 
 | | CRYSTAL STRUCTURE OF CODV IN COMPLEX WITH IL4 AT 2.55 Ang. RESOLUTION. | | Descriptor: | Antibody fragment light chain, Interleukin-4, PHOSPHATE ION, ... | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
5HCG
 
 | | CRYSTAL STRUCTURE OF FREE-CODV. | | Descriptor: | CODV heavy-chain, CODV light chain | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
1KY2
 
 | | GPPNHP-BOUND YPT7P AT 1.6 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1KY3
 
 | | GDP-BOUND YPT7P AT 1.35 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1DFE
 
 | | NMR STRUCTURE OF RIBOSOMAL PROTEIN L36 FROM THERMUS THERMOPHILUS | | Descriptor: | L36 RIBOSOMAL PROTEIN, ZINC ION | | Authors: | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | Deposit date: | 1999-11-19 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
1DGZ
 
 | | RIBOSMAL PROTEIN L36 FROM THERMUS THERMOPHILUS: NMR STRUCTURE ENSEMBLE | | Descriptor: | PROTEIN (L36 RIBOSOMAL PROTEIN), ZINC ION | | Authors: | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | Deposit date: | 1999-11-27 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
4AJW
 
 | | Discovery and Optimization of New Benzimidazole- and Benzoxazole-Pyrimidone Selective PI3KBeta Inhibitors for the Treatment of Phosphatase and TENsin homologue (PTEN)-Deficient Cancers | | Descriptor: | 2-[(1-methyl-1H-benzimidazol-2-yl)methyl]-6-morpholin-4-ylpyrimidin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Certal, V, Halley, F, Virone-Oddos, A, Delorme, C, Karlsson, A, Rak, A, Thompson, F, Filoche-Romme, B, El-Ahmad, Y, Carry, J.C, Abecassis, P.Y, Lejeune, P, Bonnevaux, H, Nicolas, J.P, Bertrand, T, Marquette, J.P, Michot, N, Benard, T, Below, P, Vade, I, Chatreaux, F, Lebourg, G, Pilorge, F, Angouillant-Boniface, O, Louboutin, A, Lengauer, C, Schio, L. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of New Benzimidazole- and Benzoxazole-Pyrimidone Selective Pi3Kbeta Inhibitors for the Treatment of Phosphatase and Tensin Homologue (Pten)-Deficient Cancers.
J.Med.Chem., 55, 2012
|
|
4BFR
 
 | | Discovery and Optimization of Pyrimidone Indoline Amide PI3Kbeta Inhibitors for the Treatment of Phosphatase and TENsin homologue (PTEN)-Deficient Cancers | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC S SUBUNIT BETA ISOFORM | | Authors: | Certal, V, Carry, J.C, Halley, F, Virone-Oddos, A, Thompson, F, Filoche-Romme, B, El-Ahmad, Y, Karlsson, A, Charrier, V, Delorme, C, Rak, A, Abecassis, P.Y, Amara, C, Vincent, L, Bonnevaux, H, Nicolas, J.P, Mathieu, M, Bertrand, T, Marquette, J.P, Michot, N, Benard, T, Perrin, M.A, Perron, S, Monget, S, Gruss-Leleu, F, Doerflinger, G, Guizani, H, Brollo, M, Delbarre, L, Bertin, L, Richepin, P, Loyau, V, Garcia-Echeverria, C, Lengauer, C, Schio, L. | | Deposit date: | 2013-03-22 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Pyrimidone Indoline Amide Pi3Kbeta Inhibitors for the Treatment of Phosphatase and Tensin Homologue (Pten)-Deficient Cancers.
J.Med.Chem., 57, 2014
|
|
1H7Z
 
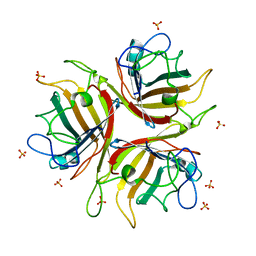 | | Adenovirus Ad3 fibre head | | Descriptor: | ADENOVIRUS FIBRE PROTEIN, SULFATE ION | | Authors: | Durmort, C, Stehlin, C, Schoehn, G, Mitraki, A, Drouet, E, Cusack, S, Burmeister, W.P. | | Deposit date: | 2001-01-21 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Fiber Head of Ad3, a Non-Car-Binding Serotype of Adenovirus
Virology, 285, 2001
|
|
6MFQ
 
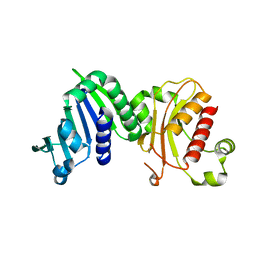 | | Crystal structure of a PMS2 variant | | Descriptor: | Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of two variants of uncertain significance in the PMS2 gene.
Hum. Mutat., 40, 2019
|
|
1QBA
 
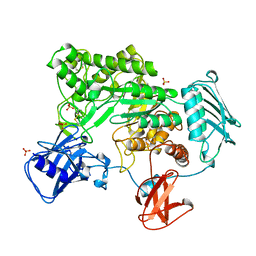 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QG0
 
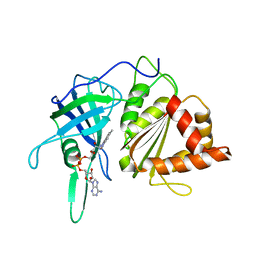 | | WILD-TYPE PEA FNR | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE) | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
