2PMV
 
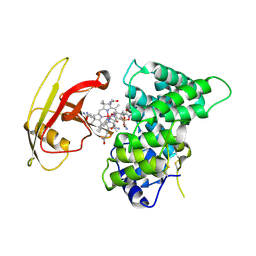 | | Crystal Structure of Human Intrinsic Factor- Cobalamin Complex at 2.6 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALAMIN, Gastric intrinsic factor | | Authors: | Mathews, F.S, Gordon, M.M, Chen, Z, Rajashankar, K.R, Ealick, S.E, Alpers, D.H, Sukumar, N. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-A resolution
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3VRS
 
 | | Crystal structure of fluoride riboswitch, soaked in Mn2+ | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MANGANESE (II) ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4R0D
 
 | | Crystal structure of a eukaryotic group II intron lariat | | Descriptor: | GROUP IIB INTRON LARIAT, IRIDIUM HEXAMMINE ION, LIGATED EXONS, ... | | Authors: | Robart, A.R, Chan, R.T, Peters, J.K, Rajashankar, K.R, Toor, N. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Crystal structure of a eukaryotic group II intron lariat.
Nature, 514, 2014
|
|
4RGF
 
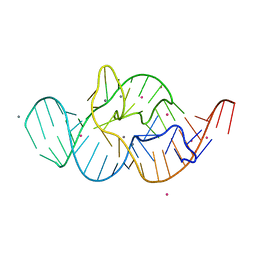 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
5KQR
 
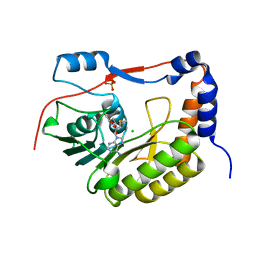 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine | | Descriptor: | CHLORIDE ION, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Jain, R, Coloma, J, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
5KK5
 
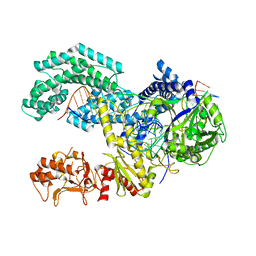 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | Authors: | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | Deposit date: | 2016-06-21 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.289 Å) | | Cite: | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
5L2X
 
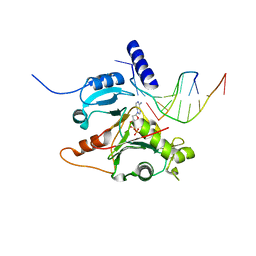 | | Crystal structure of human PrimPol ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*GP*TP*AP*GP*CP*(DDG))-3'), ... | | Authors: | Rechkoblit, O, Gupta, Y.K, Malik, R, Rajashankar, K.R, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of human PrimPol, a DNA polymerase with primase activity.
Sci Adv, 2, 2016
|
|
5KQS
 
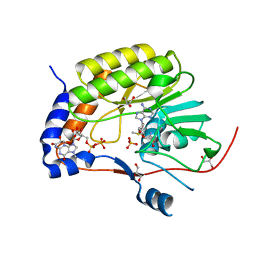 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine and 7-methyl-guanosine-5'-diphosphate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ACETATE ION, GLYCEROL, ... | | Authors: | Coloma, J, Jain, R, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
4ENC
 
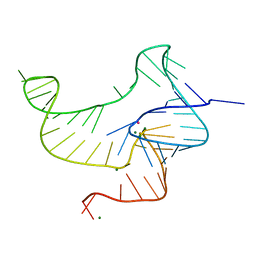 | | Crystal structure of fluoride riboswitch | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
3D12
 
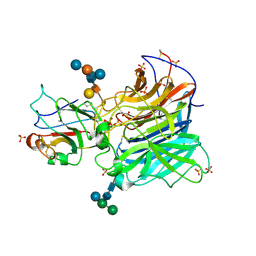 | | Crystal Structures of Nipah Virus G Attachment Glycoprotein in Complex with its Receptor Ephrin-B3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-B3, ... | | Authors: | Xu, K, Rajashankar, K.R, Chan, Y.P, Himanen, P, Broder, C.C, Nikolov, D.B. | | Deposit date: | 2008-05-02 | | Release date: | 2008-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D36
 
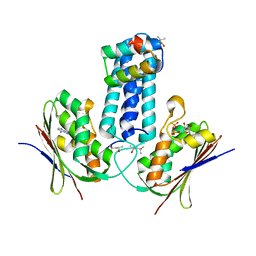 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
4ENB
 
 | | Crystal structure of fluoride riboswitch, bound to Iridium | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4EN5
 
 | | Crystal structure of fluoride riboswitch, Tl-Acetate soaked | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENA
 
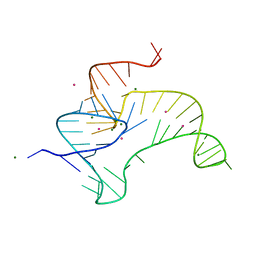 | | Crystal structure of fluoride riboswitch, soaked in Cs+ | | Descriptor: | CESIUM ION, FLUORIDE ION, Fluoride riboswitch, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
3D11
 
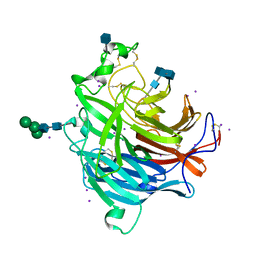 | | Crystal Structures of the Nipah G Attachment Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-neuraminidase, ... | | Authors: | Xu, K, Rajashankar, K.R, Chan, Y.P, Himanen, P, Broder, C.C, Nikolov, D.B. | | Deposit date: | 2008-05-02 | | Release date: | 2008-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5U34
 
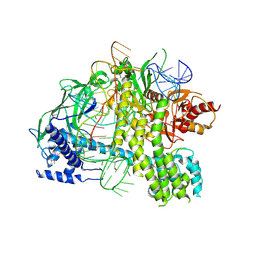 | | Crystal structure of AacC2c1-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, sgRNA | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.255 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5U30
 
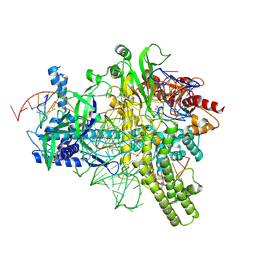 | | Crystal structure of AacC2c1-sgRNA-extended target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5U33
 
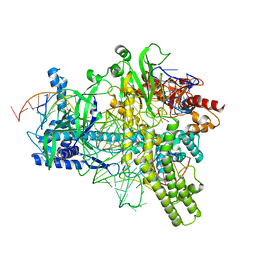 | | Crystal structure of AacC2c1-sgRNA-extended non-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5U31
 
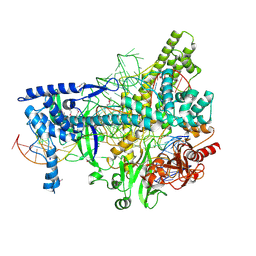 | | Crystal structure of AacC2c1-sgRNA-8mer substrate DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5UQZ
 
 | | Structural Analysis of the Glucan Binding Protein C of Streptococcus mutans Provides Evidence that it Mediates both Sucrose-Independent and -Dependent Adherence | | Descriptor: | CALCIUM ION, Glucan-binding protein C, GbpC | | Authors: | Larson, M.R, Purushotham, S, Mieher, J, Wu, R, Rajashankar, K.R, Wu, H, Deivanayagam, C. | | Deposit date: | 2017-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Glucan Binding Protein C of Streptococcus mutans Mediates both Sucrose-Independent and Sucrose-Dependent Adherence.
Infect. Immun., 86, 2018
|
|
4L3N
 
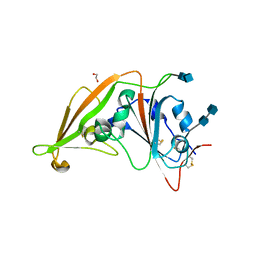 | | Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Chen, Y, Rajashankar, K.R, Yang, Y, Agnihothram, S.S, Liu, C, Lin, Y.-L, Baric, R.S, Li, F. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the receptor-binding domain from newly emerged middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
6CIH
 
 | | Crystal structure of a group II intron lariat in the post-catalytic state | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*AP*C*-3'), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
6D9Z
 
 | | Structure of CysZ, a sulfate permease from Pseudomonas Denitrificans | | Descriptor: | Sulfate transporter CysZ, octyl beta-D-glucopyranoside | | Authors: | Sanghai, Z.A, Clarke, O.B, Liu, Q, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-30 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4021318 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
6CHR
 
 | | Crystal structure of a group II intron lariat with an intact 3' splice site (pre-2s state) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*A)-3'), RNA (621-MER), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
