3IWT
 
 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2EA1
 
 | | Crystal structure of Ribonuclease I from Escherichia coli COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE | | Descriptor: | GUANYLYL-2',5'-PHOSPHOGUANOSINE, Ribonuclease I | | Authors: | Zhou, K, Pan, J, Padmanabhan, S, Lim, R.W, Lim, L.W. | | Deposit date: | 2007-01-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Ribonuclease I from Escherichia Coli Complexed with Guanylyl-2(Prime),5(Prime)-Guanosine at 1.80 Angstroms Resolution
To be Published
|
|
7PLS
 
 | | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tissue alpha-L-fucosidase | | Authors: | Armstrong, Z, Meek, R.W, Wu, L, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis.
Structure, 30, 2022
|
|
7PM4
 
 | | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Tissue alpha-L-fucosidase | | Authors: | Armstrong, Z, Meek, R.W, Wu, L, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis.
Structure, 30, 2022
|
|
7QQH
 
 | | Crystal structure of MYORG (D520N) in complex with Gal-a1,4-Glc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
7QQG
 
 | | Crystal structure of MYORG bound to 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
7QQF
 
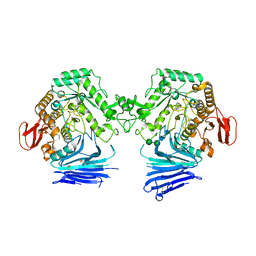 | | Crystal structure of unliganded MYORG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
3D1Z
 
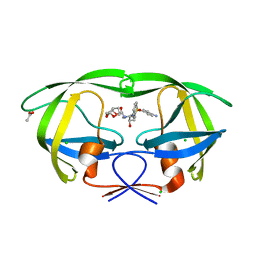 | | Crystal structure of HIV-1 mutant I54M and inhibitor DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Liu, F, Kovalesky, A.Y, Tie, Y, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2008-05-07 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3T0B
 
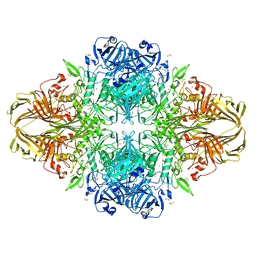 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2Q
 
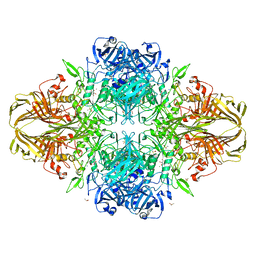 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T08
 
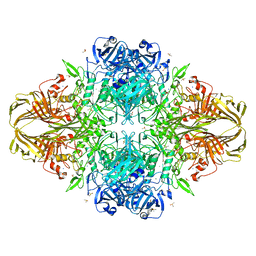 | | E. coli (LacZ) beta-galactosidase (S796A) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0D
 
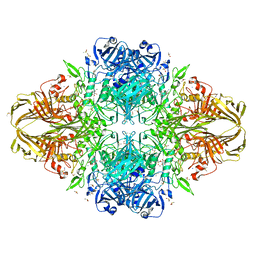 | | E.coli (lacZ) beta-galactosidase (S796T) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ser-796 of Beta-galactosidase (E. coli) plays a Key role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3SEP
 
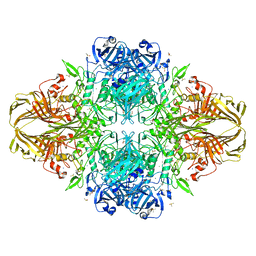 | | E. coli (lacZ) beta-galactosidase (S796A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3CB0
 
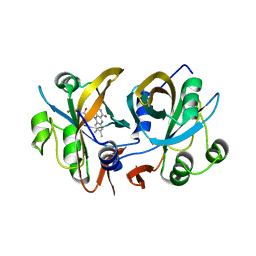 | | CobR | | Descriptor: | 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE, FLAVIN MONONUCLEOTIDE | | Authors: | Lawrence, A.D, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis
J.Biol.Chem., 283, 2008
|
|
3CCW
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[4-(benzylcarbamoyl)-2-(4-fluorophenyl)-5-(1-methylethyl)-1H-imidazol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CD0
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{2-[(4-fluorobenzyl)carbamoyl]-4-(4-fluorophenyl)-1-(1-methylethyl)-1H-imidazol-5-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CD7
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CDB
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{3-[(4-carbamoylphenyl)sulfamoyl]-4,5-bis(4-fluorophenyl)-2-(1-methylethyl)-1H-pyrrol-1-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CDA
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{3-(4-fluorophenyl)-1-(1-methylethyl)-4-phenyl-5-[(4-sulfamoylphenyl)carbamoyl]-1H-pyrrol-2-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3T09
 
 | | E. coli (LacZ) beta-galactosidase (S796A) galactonolactone complex | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
2FAI
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-2M and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2S,5S,9R)-5-(HYDROXYMETHYL)-8,9-DIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
3T2P
 
 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
2ETE
 
 | | Recombinant oxalate oxidase in complex with glycolate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYOXYLIC ACID, MANGANESE (II) ION, ... | | Authors: | Opaleye, O, Rose, R.-S, Whittaker, M.M, Woo, E.-J, Whittaker, J.W, Pickersgill, R.W. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and spectroscopic studies shed light on the mechanism of oxalate oxidase
J.Biol.Chem., 281, 2006
|
|
3DDX
 
 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | Descriptor: | Major capsid protein | | Authors: | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | Deposit date: | 2008-06-06 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY | | Cite: | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
3CCT
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-4-[(2-hydroxyphenyl)carbamoyl]-5-(1-methylethyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
