4I34
 
 | | Crystal Structure of W-W-W ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1218 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4Q2Z
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | Descriptor: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | Authors: | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | Deposit date: | 2014-04-10 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3AXD
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
1K74
 
 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K7L
 
 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4XLY
 
 | | The complex structure of KS-D75C with substrate CPP | | Descriptor: | (2E)-3-methyl-5-[(1R,4aR,8aR)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
1KE8
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE | | Descriptor: | 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE6
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KKQ
 
 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
4XLX
 
 | | Crystal structure of BjKS from Bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
1KE9
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
4XWF
 
 | |
1KE7
 
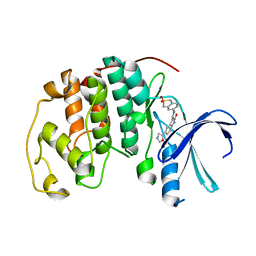 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
4XW7
 
 | |
1KE5
 
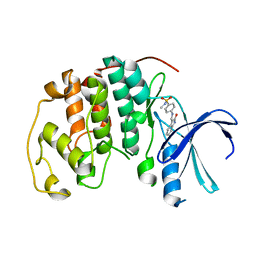 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
4XP7
 
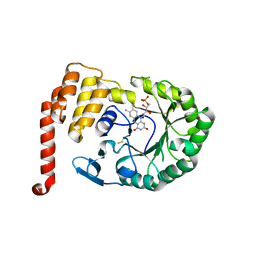 | | Crystal structure of Human tRNA dihydrouridine synthase 2 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Whelan, F, Jenkins, H.T, Griffiths, S, Byrne, R.T, Dodson, E.J, Antson, A.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From bacterial to human dihydrouridine synthase: automated structure determination.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3DDW
 
 | | Crystal structure of glycogen phosphorylase complexed with an anthranilimide based inhibitor GSK055 | | Descriptor: | (2S)-{[(3-{[(2-chloro-6-methylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]amino}(phenyl)ethanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4YCO
 
 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNAPhe | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YJX
 
 | | The structure of Agrobacterium tumefaciens ClpS2 bound to L-phenylalaninamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, PHENYLALANINE AMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
3CJF
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
4Z2B
 
 | | The structure of human PDE12 residues 161-609 in complex with GSK3036342A | | Descriptor: | 1,2-ETHANEDIOL, 2',5'-phosphodiesterase 12, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
4Z0V
 
 | | The structure of human PDE12 residues 161-609 | | Descriptor: | 2',5'-phosphodiesterase 12, GLYCEROL, MAGNESIUM ION | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-26 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
3D0X
 
 | |
