7ROX
 
 | | BthTX-I complexed with inhibitor MMV | | Descriptor: | 12-methoxy-Nb-methylvoachalotine, Basic phospholipase A2 homolog bothropstoxin-I | | Authors: | Borges, R.J, De Marino, I, Uson, I, Fontes, M.R.M. | | Deposit date: | 2021-08-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of a snake venom phospholipase A 2 -like protein complexed to an inhibitor from Tabernaemontana catharinensis.
Biochimie, 206, 2023
|
|
6QQ5
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric oxide reductase subunit B, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
6QU6
 
 | | Adenovirus Serotype 26 (Ad26) in complex with sialic acid, pH4.0 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fiber, ... | | Authors: | Baker, A.T, Mundy, R.M, Parker, A.L, Rizkallah, P.J. | | Deposit date: | 2019-02-26 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor.
Sci Adv, 5, 2019
|
|
6I0O
 
 | | Structure of human IMP dehydrogenase, isoform 2, bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase 2, SULFATE ION | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity.
J. Mol. Biol., 431, 2019
|
|
6QQ6
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
8CLM
 
 | |
8CKS
 
 | | Crystal structure of Human Serum Albumin in complex with FESAN | | Descriptor: | 3,3'-commo-bis(1,2-dicarba-3-ferra-closo-dodecaborane), DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Dolot, R.M, Kaniowski, D, Ebenryter-Olbinska, K, Szczupak, P, Suwara, J, Nawrot, B.C. | | Deposit date: | 2023-02-16 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Serum Albumin in complex with FESAN
To Be Published
|
|
8CNX
 
 | | Structure of Enterovirus D68 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of EV D68 3C protease
To Be Published
|
|
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of EV D68 3C protease - to be published
To Be Published
|
|
5OJE
 
 | | Endothiapepsin with Ligand VSK-B24 | | Descriptor: | (2~{S})-2-azanyl-3-(1~{H}-indol-3-yl)-~{N}-[2-(2,4,6-trimethylphenyl)ethyl]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Gierse, R.M, Magari, F, Groves, M.R, Heine, A, Klebe, G, Hirsch, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-09-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Design and Synthesis of Bioisosteres of Acylhydrazones as Stable Inhibitors of the Aspartic Protease Endothiapepsin.
ChemMedChem, 13, 2018
|
|
7CQ2
 
 | | Crystal structure of Slx1-Slx4 | | Descriptor: | GLYCEROL, SLX4 isoform 1, Structure-specific endonuclease subunit SLX1, ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
5OQE
 
 | |
6HPF
 
 | | Structure of Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | von Freiesleben, P, Moroz, O.V, Blagova, E, Wiemann, M, Spodsberg, N, Agger, J.W, Davies, G.J, Wilson, K.S, Stalbrand, H, Meyer, A.S, Krogh, K.B.R.M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure and substrate interactions of an unusual fungal non-CBM carrying GH26 endo-beta-mannanase from Yunnania penicillata.
Sci Rep, 9, 2019
|
|
5OO3
 
 | |
7CQ3
 
 | | Crystal structure of Slx1-Slx4 | | Descriptor: | SLX4 isoform 1, SULFATE ION, Structure-specific endonuclease subunit SLX1, ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
7CQ4
 
 | | Crystal structure of Slx1-Slx4 in complex with 5'flap DNA | | Descriptor: | DNA (27-MER), DNA (5'-D(*AP*GP*GP*AP*CP*AP*TP*CP*TP*TP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*TP*AP*CP*AP*AP*CP*AP*GP*AP*T)-3'), ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
5OOZ
 
 | |
5OQ9
 
 | |
5OSM
 
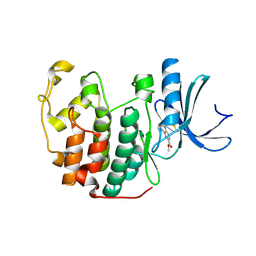 | | Cdk2(F80C, C177A) with covalent adduct at C80 | | Descriptor: | Cyclin-dependent kinase 2, methyl 1-propanoyl-3,4-dihydro-2~{H}-quinoline-6-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6I3V
 
 | | x-ray structure of the human mitochondrial PRELID1 in complex with TRIAP1 | | Descriptor: | CHLORIDE ION, MYRISTIC ACID, PRELI domain-containing protein 1, ... | | Authors: | Berry, J.L, Miliara, X, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I7S
 
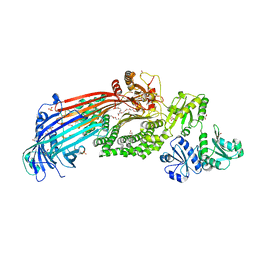 | | Microsomal triglyceride transfer protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Biterova, E, Isupov, M.N, Keegan, R.M, Lebedev, A.A, Ruddock, L.W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human microsomal triglyceride transfer protein.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XXG
 
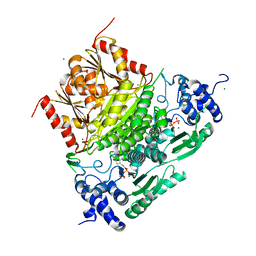 | | Structure of truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-01-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties.
Biochem.Biophys.Res.Commun., 539, 2021
|
|
4PW6
 
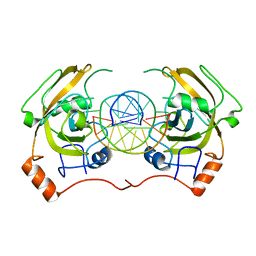 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
2YET
 
 | | Thermoascus GH61 isozyme A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, COPPER (II) ION, ... | | Authors: | Otten, H, Quinlan, R.J, Sweeney, M.D, Poulsen, J.-C.N, Johansen, K.S, Krogh, K.B.R.M, Joergensen, C.I, Tovborg, M, Anthonsen, A, Tryfona, T, Walter, C.P, Dupree, P, Xu, F, Davies, G.J, Walton, P.H, Lo Leggio, L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Insights Into the Oxidative Degradation of Cellulose by a Copper Metalloenzyme that Exploits Biomass Components.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4PW5
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex I | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
