2KAM
 
 | |
2KWA
 
 | | 1H, 13C and 15N backbone and side chain resonance assignments of the N-terminal domain of the histidine kinase inhibitor KipI from Bacillus subtilis | | Descriptor: | Kinase A inhibitor | | Authors: | Hynson, R.M.G, Kwan, A, Jacques, D.A, Mackay, J.P, Trewhella, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel Structure of an Antikinase and its Inhibitor
J.Mol.Biol., 2010
|
|
6RPU
 
 | | Structure of the ternary complex of the IMPDH enzyme from Ashbya gossypii bound to the dinucleoside polyphosphate Ap5G and GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates.
J.Biol.Chem., 294, 2019
|
|
4CV4
 
 | | PIH N-terminal domain | | Descriptor: | COBALT (II) ION, PIH1 DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
3POM
 
 | |
6EO7
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
3PR2
 
 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
6U9I
 
 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | Descriptor: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
4F57
 
 | |
6UK1
 
 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
4F58
 
 | |
2LPN
 
 | | Solution Structure of N-Terminal domain of human Conserved Dopamine Neurotrophic Factor (CDNF) | | Descriptor: | Cerebral dopamine neurotrophic factor | | Authors: | Latge, C, Cabral, K.M.S, Foguel, D, Pires, J.R.M, Almeida, M.S. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-05-22 | | Method: | SOLUTION NMR | | Cite: | (1)H-, (13)C- and (15)N-NMR assignment of the N-terminal domain of human cerebral dopamine neurotrophic factor (CDNF).
Biomol.Nmr Assign., 7, 2013
|
|
3QGZ
 
 | | Re-investigated high resolution crystal structure of histidine triad nucleotide-binding protein 1 (HINT1) from rabbit complexed with adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A, Nawrot, B, Stec, W.J. | | Deposit date: | 2011-01-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution X-ray crystal structure of rabbit histidine triad nucleotide-binding protein 1 (rHINT1) - adenosine complex at 1.10A resolution
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3QNL
 
 | | Crystal structure of PrTX-I complexed to Rosmarinic Acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ISOPROPYL ALCOHOL, ... | | Authors: | dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Functional Studies of a Bothropic Myotoxin Complexed to Rosmarinic Acid: New Insights into Lys49-PLA(2) Inhibition.
Plos One, 6, 2011
|
|
2AX2
 
 | | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Budayova-Spano, M, Fisher, S.Z, Dauvergne, M.T, Silverman, D.N, Myles, D.A.A, McKenna, R.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2ID9
 
 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID7
 
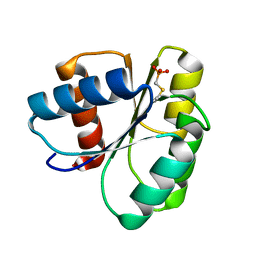 | | 1.75 A Structure of T87I Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
6B5A
 
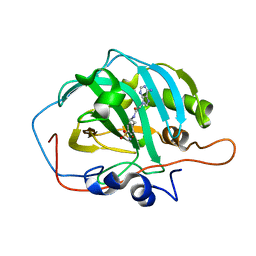 | |
6B80
 
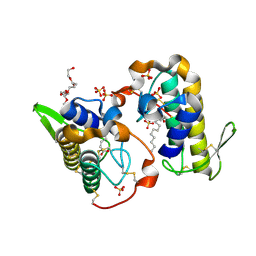 | | Crystal structure of myotoxin II from Bothrops moojeni complexed to myristic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog 2, MYRISTIC ACID, ... | | Authors: | Salvador, G.H.M, dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural evidence for a fatty acid-independent myotoxic mechanism for a phospholipase A2-like toxin.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
2DIJ
 
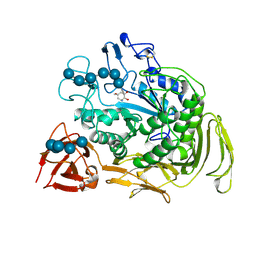 | | COMPLEX OF A Y195F MUTANT CGTASE FROM B. CIRCULANS STRAIN 251 COMPLEXED WITH A MALTONONAOSE INHIBITOR AT PH 9.8 OBTAINED AFTER SOAKING THE CRYSTAL WITH ACARBOSE AND MALTOHEXAOSE | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Knegtel, R.M.A, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 1998-05-27 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity.
Biochemistry, 35, 1996
|
|
6B59
 
 | |
6B81
 
 | |
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
6C5Z
 
 | | Human UDP-Glucose Dehydrogenase A225L substitutuion with UDP-glucose and NADH bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Gross, P.G, Sidlo, A.M, Walsh, R.M, Peeples, W.B, Wood, Z.A. | | Deposit date: | 2018-01-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The A225L Substitution of hUGDH alters structure and function
To Be Published
|
|
4JO3
 
 | |
